Summary
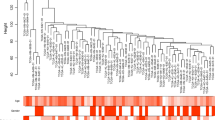
Renal cancer is a common genitourinary malignance, of which clear cell renal cell carcinoma (ccRCC) has high aggressiveness and leads to most cancer-related deaths. Identification of sensitive and reliable biomarkers for predicting tumorigenesis and progression has great significance in guiding the diagnosis and treatment of ccRCC. Here, we identified 2397 common differentially expressed genes (DEGs) using paired normal and tumor ccRCC tissues from GSE53757 and The Cancer Genome Atlas (TCGA). Then, we performed weighted gene co-expression network analysis and protein-protein interaction network analysis, 17 candidate hub genes were identified. These candidate hub genes were further validated in GSE36895 and Oncomine database and 14 real hub genes were identified. All the hub genes were up-regulated and significantly positively correlated with pathological stage and histologic grade of ccRCC. Survival analysis showed that the higher expression level of each hub gene tended to predict a worse clinical outcome. ROC analysis showed that all the hub genes can accurately distinguish between tumor and normal samples, and between early stage and advanced stage ccRCC. Moreover, all the hub genes were positively associated with distant metastasis, lymph node infiltration, tumor recurrence and the expression of MKi67, suggesting these genes might promote tumor proliferation, invasion and metastasis. Furthermore, the functional annotation demonstrated that most genes were enriched in cell-cycle related biological function. In summary, our study identified 14 potential biomarkers for predicting tumorigenesis and progression, which might contribute to early diagnosis, prognosis prediction and therapeutic intervention.
Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin, 2019,69(1):7–34
Ljungberg B, Bensalah K, Canfield S, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol, 2015,67(5):913–924
Pierorazio PM, Johnson MH, Patel HD, et al. Management of Renal Masses and Localized Renal Cancer: Systematic Review and Meta-Analysis. J Urol, 2016,196(4):989–999
Bazzi WM, Sjoberg DD, Feuerstein MA, et al. Long-Term Survival Rates after Resection for Locally Advanced Kidney Cancer: Memorial Sloan Kettering Cancer Center 1989 to 2012 Experience. J Urol, 2015,193(6):1911–1916
Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers, 2017,3:17009
Zhao E, Li L, Zhang W, et al. Comprehensive characterization of immune- and inflammation-associated biomarkers based on multi-omics integration in kidney renal clear cell carcinoma. J Transl Med, 2019,17(1):177
Guan LY, Tan JF, Li H, et al. Biomarker identification in clear cell renal cell carcinoma based on miRNA-seq and digital gene expression-seq data. Gene, 2018,647:205–212
Tavazoie S, Hughes JD, Campbell MJ, et al. Systematic determination of genetic network architecture. Nat Genet, 1999,22(3):281–285
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 2008,9:559
Clarke C, Madden SF, Doolan P, et al. Correlating transcriptional networks to breast cancer survival: a large-scale coexpression analysis. Carcinogenesis, 2013,34(10):2300–2308
Wang L, Tang H, Thayanithy V, et al. Gene networks and microRNAs implicated in aggressive prostate cancer. Cancer Res, 2009,69(24):9490–9497
Gu HY, Yang M, Guo J, et al. Identification of the Biomarkers and Pathological Process of Osteoarthritis: Weighted Gene Co-expression Network Analysis. Front Physiol, 2019,10:275
Chen L, Yuan L, Qian K, et al. Identification of Biomarkers Associated With Pathological Stage and Prognosis of Clear Cell Renal Cell Carcinoma by Coexpression Network Analysis. Front Physiol, 2018,9:399
Ficarra V, Schips L, Guille F, et al. Multiinstitutional European validation of the 2002 TNM staging system in conventional and papillary localized renal cell carcinoma. Cancer, 2005,104(5):968–974
Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol, 2008,4(8):e1000117
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc, 2009,4(1):44–57
Goel MK, Khanna P, Kishore J. Understanding survival analysis: Kaplan-Meier estimate. Int J Ayurveda Res, 2010,1(4):274–278
Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res, 2017,45(W1):W98–W102
Sing T, Sander O, Beerenwinkel N, et al. ROCR: visualizing classifier performance in R. Bioinformatics, 2005,21(20):3940–3941
Gayed BA, Youssef RF, Bagrodia A, et al. Ki67 is an independent predictor of oncological outcomes in patients with localized clear-cell renal cell carcinoma. BJU Int, 2014,113(4):668–673
Tilki D, Nguyen HG, Dall’Era MA, et al. Impact of histologic subtype on cancer-specific survival in patients with renal cell carcinoma and tumor thrombus. Eur Urol, 2014,66(3):577–583
Gospodarowicz MK, Miller D, Groome PA, et al. The process for continuous improvement of the TNM classification. Cancer, 2004,100(1):1–5
Xu J, Latif S, Wei S. Metastatic renal cell carcinoma presenting as gastric polyps: A case report and review of the literature. Int J Surg Case Rep, 2012,3(12):601–604
Frank I, Blute ML, Cheville JC, et al. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol, 2002,168(6):2395–400
Chen L, Yuan LS, Wang YZ, et al. Co-expression network analysis identified FCER1G in association with progression and prognosis in human clear cell renal cell carcinoma. Int J Biol Sci, 2017,13(11):1361–1372
Yuan L, Chen L, Qian K, et al. Co-expression network analysis identified six hub genes in association with progression and prognosis in human clear cell renal cell carcinoma (ccRCC). Genom Data, 2017,14:132–140
Wang Y, Chen L, Wang G, et al. Fifteen hub genes associated with progression and prognosis of clear cell renal cell carcinoma identified by coexpression analysis. J Cell Physiol, 2019,234(7):10225–10237
Dachineni R, Ai GQ, Kumar DR, et al. Cyclin A2 and CDK2 as Novel Targets of Aspirin and Salicylic Acid: A Potential Role in Cancer Prevention. Mol Cancer Res, 2016,14(3):241–252
Lei CY, Wang W, Zhu YT, et al. The decrease of cyclin B2 expression inhibits invasion and metastasis of bladder cancer. Urol Oncol, 2016,34(5):237.e1–10
Mo ML, Chen Z, Li J, et al. Use of serum circulating CCNB2 in cancer surveillance. Int J Biol Markers, 2010,25(4):236–242
Duxbury MS, Whang EE. RRM2 induces NF-kappaB-dependent MMP-9 activation and enhances cellular invasiveness. Biochem Biophys Res Commun, 2007,354(1):190–196
Zhang K, Hu S, Wu J, et al. Overexpression of RRM2 decreases thrombspondin-1 and increases VEGF production in human cancer cells in vitro and in vivo: implication of RRM2 in angiogenesis. Mol Cancer, 2009,8:11
Liu X, Zhou B, Xue L, et al. Ribonucleotide reductase subunits M2 and p53R2 are potential biomarkers for metastasis of colon cancer. Clin Colorectal Cancer, 2007,6(5):374–381
Gartel AL. FOXM1 in Cancer: Interactions and Vulnerabilities. Cancer Res, 2017,77(12):3135–3139
Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta, 2012,1819(1):28–37
Guo L, Ding Z, Huang N, et al. Forkhead Box M1 positively regulates UBE2C and protects glioma cells from autophagic death. Cell Cycle, 2017,16(18):1705–1718
Hu GH, Yan ZW, Zhang C, et al. FOXM1 promotes hepatocellular carcinoma progression by regulating KIF4A expression. J Exp Clin Cancer Res, 2019,38
Khongkow P, Gomes AR, Gong C, et al. Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene, 2016,35(8):990–1002
Kalimutho M, Sinha D, Jeffery J, et al. CEP55 is a determinant of cell fate during perturbed mitosis in breast cancer. EMBO Mol Med, 2018,10(9)
Li Y, Zhang ZF, Chen JD, et al. VX680/MK-0457, a potent and selective Aurora kinase inhibitor, targets both tumor and endothelial cells in clear cell renal cell carcinoma. Am J Transl Res, 2010,2(3):296–308
van Gijn SE, Wierenga E, van den Tempel N, et al. TPX2/Aurora kinase A signaling as a potential therapeutic target in genomically unstable cancer cells. Oncogene, 2019,38(6):852–867
Hu P, Chen X, Sun J, et al. siRNA-mediated knockdown against NUF2 suppresses pancreatic cancer proliferation in vitro and in vivo. Biosci Rep, 2015,35(1):e00170
Fu HL, Shao L. Silencing of NUF2 inhibits proliferation of human osteosarcoma Saos-2 cells. Eur Rev Med Pharmacol Sci, 2016,20(6):1071–1079
Ghaffari K, Hashemi M, Ebrahimi E, et al. BIRC5 Genomic Copy Number Variation in Early-Onset Breast Cancer. Iran Biomed J, 2016,20(4):241–245
Ren Q, Jin B. The clinical value and biological function of PTTG1 in colorectal cancer. Biomed Pharmacother, 2017,89:108–115
Zhang Q, Su RX, Shan C, et al. Non-SMC Condensin I Complex, Subunit G (NCAPG) is a Novel Mitotic Gene Required for Hepatocellular Cancer Cell Proliferation and Migration. Oncol Res, 2018,26(2):269–276
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that there is no conflict of interest with any financial organization or corporation or individual that can inappropriately influence this work.
Additional information
This work was supported by grants from the National Natural Science Foundation of China (No. 81270354) and Natural Science for Youth Foundation (No. 81300213).
Rights and permissions
About this article
Cite this article
Chen, Jy., Sun, Y., Qiao, N. et al. Co-expression Network Analysis Identifies Fourteen Hub Genes Associated with Prognosis in Clear Cell Renal Cell Carcinoma. CURR MED SCI 40, 773–785 (2020). https://doi.org/10.1007/s11596-020-2245-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11596-020-2245-6