Summary
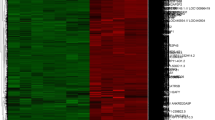
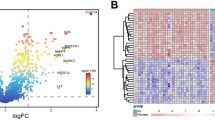
Systemic sclerosis (SSc) is a highly heterogeneous autoimmune disease with a high mortality rate. However, the cellular and molecular mechanisms of SSc remain unclear. Here, we identified the key hub genes and microRNAs (miRNAs) that modulate the occurrence and development of SSc. We downloaded the microarray dataset GSE95065 from the Gene Expression Omnibus (GEO) database and then analyzed the data by using GEO2R. The Database for Annotation, Visualization and Integrated Discovery (DAVID) was used for functional pathway enrichment analyses of differentially expressed genes (DEGs), and Cytoscape software was used to generate the protein-protein interaction (PPI) network. In addition, OmicsNet was used to predict the miRNAs for the hub genes of SSc. As a result, 783 DEGs were identified, of which 770 genes (142 up-regulated genes and 628 down-regulated genes) were matched to the genes in SSc skin samples. Gene Ontology (GO) analyses by DAVID indicated that the up-regulated genes were mainly involved in immune response, and the down-regulated genes were greatly enriched in glycinergic synaptic transmission. In the PPI network, 22 nodes were selected as key genes, including several members of the chemokine family. Furthermore, after uploading these key genes to the OmicsNet tool, we found that hsa-miR-26b-5p might target CXCL9 and CXCL13. Moreover, we demonstrated that the hsa-miR-26b-5p inhibitor might inhibit fibrosis in TGF-β-activated fibroblasts, which would be a promising target for SSc therapy.
Similar content being viewed by others
References
Barnes J, Mayes MD. Epidemiology of systemic sclerosis: incidence, prevalence, survival, risk factors, malignancy, and environmental triggers. Curr Opin Rheumatol, 2012,24(2):165–170
Denton CP. Advances in pathogenesis and treatment of systemic sclerosis. Clin Med (Lond), 2016,16(1):55–60
Denton CP. Systemic sclerosis: from pathogenesis to targeted therapy. Clin Exp Rheumatol, 2015,33(4 Suppl 92):S3–S7
Zuo X, Zhang L, Luo H, et al. Systematic approach to understanding the pathogenesis of systemic sclerosis. Clin Genet, 2017,92(4):365–371
Sing T, Jinnin M, Yamane K, et al. microRNA-92a expression in the sera and dermal fibroblasts increases in patients with scleroderma. Rheumatology (Oxford), 2012,51(9):1550–1556
Zhu H, Luo H, Li Y, et al. MicroRNA-21 in scleroderma fibrosis and its function in TGF-beta-regulated fibrosis-related genes expression. J Clin Immunol, 2013,33(6):1100–1109
Miao CG, Xiong YY, Yu H, et al. Critical roles of microRNAs in the pathogenesis of systemic sclerosis: New advances, challenges and potential directions. Int Immunopharmacol, 2015,28(1):626–633
Clough E, Barrett T. The Gene Expression Omnibus Database. Methods Mol Biol, 2016,1418:93–110
Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc, 2015,10(6):823–844
Gene Ontology C. The Gene Ontology (GO) project in 2006. Nucleic Acids Res, 2006,34(Database issue):D322–326
Lewis SE. The Vision and Challenges of the Gene Ontology. Methods Mol Biol, 2017,1446:291–302
Kanehisa M, Sato Y, Kawashima M, et al. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res, 2016,44(D1):D457–462
Huang DW, Sherman BT, Tan Q, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res, 2007,35(Web Server issue):W169–175
Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res, 2015,43(Database issue):D447–452
Su G, Morris JH, Demchak B, et al. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics, 2014,47:8.13.1–24
Zhou G, Xia J. OmicsNet: a web-based tool for creation and visual analysis of biological networks in 3D space. Nucleic Acids Res, 2018,46(W1):W514–W522
Elia G, Guglielmi G. CXCL9 chemokine in ulcerative colitis. Clin Ter, 2018,169(5):e235–e241
Ding Q, Lu P, Xia Y, et al. CXCL9: evidence and contradictions for its role in tumor progression. Cancer Med, 2016,5(11):3246–3259
Duarte GV, Boeira V, Correia T, et al. Osteopontin, CCL5 and CXCL9 are independently associated with psoriasis, regardless of the presence of obesity. Cytokine, 2015,74(2):287–292
Su R, Nguyen ML, Agarwal MR, et al. Interferon-inducible chemokines reflect severity and progression in sarcoidosis. Respir Res, 2013,14:121
Wenzel J, Tuting T. Identification of type I interferon-associated inflammation in the pathogenesis of cutaneous lupus erythematosus opens up options for novel therapeutic approaches. Exp Dermatol, 2007,16(5):454–463
O’Brien JC, Rainwater YB, Malviya N, et al. Transcriptional and Cytokine Profiles Identify CXCL9 as a Biomarker of Disease Activity in Morphea. J Invest Dermatol, 2017,137(8):1663–1670
Rabquer BJ, Tsou PS, Hou Y, et al. Dysregulated expression of MIG/CXCL9, IP-10/CXCL10 and CXCL16 and their receptors in systemic sclerosis. Arthritis Res Ther, 2011,13(1):R18
Hasegawa M, Fujimoto M, Matsushita T, et al. Serum chemokine and cytokine levels as indicators of disease activity in patients with systemic sclerosis. Clin Rheumatol, 2011,30(2):231–237
Hasegawa M, Asano Y, Endo H, et al. Serum chemokine levels as prognostic markers in patients with early systemic sclerosis: a multicenter, prospective, observational study. Mod Rheumatol, 2013,23(6):1076–1084
Kuo PT, Zeng Z, Salim N, et al. The Role of CXCR3 and Its Chemokine Ligands in Skin Disease and Cancer. Front Med (Lausanne), 2018,5:271
Fazilleau N, Mark L, McHeyzer-Williams LJ, et al. Follicular helper T cells: lineage and location. Immunity, 2009,30(3):324–335
Lim HW, Hillsamer P, Kim CH. Regulatory T cells can migrate to follicles upon T cell activation and suppress GC-Th cells and GC-Th cell-driven B cell responses. J Clin Invest, 2004,114(11):1640–1649
Shi K, Hayashida K, Kaneko M, et al. Lymphoid Chemokine B Cell-Attracting Chemokine-1 (CXCL13) Is Expressed in Germinal Center of Ectopic Lymphoid Follicles within the Synovium of Chronic Arthritis Patients. J Immunol, 2001,166(1):650–655
Ishikawa S, Sato T, Abe M, et al. Aberrant high expression of B lymphocyte chemokine (BLC/CXCL13) by C11b+CD11c+ dendritic cells in murine lupus and preferential chemotaxis of B1 cells towards BLC. J Exp Med, 2001,193(12):1393–1402
Kramer JM, Klimatcheva E, Rothstein TL. CXCL13 is elevated in Sjogren’s syndrome in mice and humans and is implicated in disease pathogenesis. J Leukoc Biol, 2013,94(5):1079–1089
Wutte N, Kovacs G, Berghold A, et al. CXCL13 and B-cell activating factor as putative biomarkers in systemic sclerosis. Br J Dermatol, 2013,169(3):723–725
Taniguchi T, Miyagawa T, Toyama S, et al. CXCL13 produced by macrophages due to Fli1 deficiency may contribute to the development of tissue fibrosis, vasculopathy and immune activation in systemic sclerosis. Exp Dermatol, 2018,27(9):1030–1037
Ammirante M, Shalapour S, Kang Y, et al. Tissue injury and hypoxia promote malignant progression of prostate cancer by inducing CXCL13 expression in tumor myofibroblasts. Proc Natl Acad Sci USA, 2014,111(41):14776–14781
Wei Y, Lin C, Li H, et al. CXCL13 expression is prognostic and predictive for postoperative adjuvant chemotherapy benefit in patients with gastric cancer. Cancer Immunol Immunother, 2018,67(2):261–269
Zhu Z, Zhang X, Guo H, et al. CXCL13-CXCR5 axis promotes the growth and invasion of colon cancer cells via PI3K/AKT pathway. Mol Cell Biochem, 2015,400(1-2):287–295
Cai Y, Yu X, Hu S, et al. A brief review on the mechanisms of miRNA regulation. Genomics, Proteomics Bioinformatics, 2009,7(4):147–154
Miao CG, Xiong YY, Yu H, et al. Critical roles of microRNAs in the pathogenesis of systemic sclerosis: New advances, challenges and potential directions. Int Immunopharmacol, 2015,28(1):626–633
Jia CM, Tian YY, Quan LN, et al. miR-26b-5p suppresses proliferation and promotes apoptosis in multiple myeloma cells by targeting JAG1. Pathol Res Pract, 2018,214(9):1388–1394
Fan F, Lu J, Yu W, et al. MicroRNA-26b-5p regulates cell proliferation, invasion and metastasis in human intrahepatic cholangiocarcinoma by targeting S100A7. Oncol Lett, 2018,15(1):386–392
Tang CM, Zhang M, Huang L, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep, 2017,7:40342
Chouri E, Servaas NH, Bekker CPJ, et al. Serum microRNA screening and functional studies reveal miR-483-5p as a potential driver of fibrosis in systemic sclerosis. J Autoimmun, 2018,89:162–170
Richmond A. Nf-kappa B, chemokine gene transcription and tumour growth. Nat Rev Immunol, 2002,2(9):664–674
Author information
Authors and Affiliations
Corresponding author
Additional information
This project was supported by National Natural Science Foundation of China (No. 81472886).
Conflict of Interest Statement
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Sun, Yh., Xie, M., Wu, Sd. et al. Identification and Interaction Analysis of Key Genes and MicroRNAs in Systemic Sclerosis by Bioinformatics Approaches. CURR MED SCI 39, 645–652 (2019). https://doi.org/10.1007/s11596-019-2086-3
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11596-019-2086-3