Abstract
Of the nearly 300 species of the phylum Glomeromycota comprising arbuscular mycorrhizal fungi (AMF), only 24 were originally described to form glomoid spores in unorganized sporocarps with a peridium and a gleba, in which the spores are distributed randomly. However, the natural (molecular) phylogeny of most of these species remains unknown. We found unorganized sporocarps of two fungi-producing glomoid spores: one in the Amazonian forest in Brazil (tropical forest) and the second in a forest of Poland (temperate forest). The unique spore morphology of the two fungi suggested that they are undescribed species. Subsequent phylogenetic analyses of sequences of the small subunit–internal transcribed spacer–large subunit nrDNA region and the RPB1 gene confirmed this assumption and placed the Brazilian fungus in a separate clade at the rank of genus, very strongly divergent from its sister clade representing the genus Glomus sensu stricto in the family Glomeraceae (order Glomerales). The Polish fungus was accommodated in a sister clade to a clade grouping sequences of Diversispora epigaea, a fungus that also occasionally produces spores in sporocarps, belonging in the Diversisporaceae (Diversisporales). Consequently, the Brazilian fungus was here described as the new genus and new species Sclerocarpum gen. nov. and S. amazonicum sp. nov., respectively. The Polish fungus was described as D. sporocarpia sp. nov. In addition, the supposed reasons for the low representation of sporocarpic species in the Glomeromycota were discussed and the known distribution of sporocarp-producing Glomeromycota was outlined.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Of the nearly 300 species of the phylum Glomeromycota C. Walker & A. Schüßler (Schüßler et al. 2001; Tedersoo et al. 2018; Turrini et al. 2018) comprising arbuscular mycorrhizal fungi (AMF), only 57 were originally described to form spores at the tip of sporogenous hyphae in epigeous or hypogeous sporocarps, i.e., fruit bodies, with or without a peridium and with a gleba. The peridium is a network of hyphae that partially or completely covers the gleba, which is composed of spores embedded in interwoven hyphae (Gerdemann and Trappe 1974; Morton 1988). Sporocarps that have highly ordered spores, usually distributed side-by-side in a single layer around a central hyphal plexus, are called organized sporocarps and within the Glomeromycota are obligatorily produced mainly by species of the genus Sclerocystis Berk. & Broome (Morton 1988). Sporocarps with spores randomly distributed in the gleba are called “unorganized sporocarps.” However, the formation of unorganized sporocarps is not always a stable character because some species that form such sporocarps, as for example Glomus macrocarpum Tul. & C. Tul., also produced spores in more- or less-compact naked clusters (without a glebal hyphae and a peridium) and/or single naked spores (Berch and Fortin 1983). According to Morton (1988), the formation of spores in naked clusters and singly by sporocarpic species probably is associated with undetermined conditions functioning in different geographical locations.
All current Glomeromycota species that produced unorganized sporocarps with glomoid spores, i.e., similar to those of G. macrocarpum, were originally described in the genus Glomus Tul. & C. Tul. (Gerdemann and Trappe 1974; Redecker et al. 2007), which currently belongs to the family Glomeraceae Piroz. & Dalpé in the order Glomerales J.B. Morton & Benny (Morton and Benny 1990; Tedersoo et al. 2018). However, molecular phylogenetic analyses placed, for example, G. fulvum (Berk. & Broome) Trappe & Gerd. and G. megalocarpum D. Redecker in a separate clade at the rank of genus next to Diversispora spurca (C.M. Pfeiff., C. Walker & Bloss) C. Walker & A. Schüßler (formerly G. spurcum C.M. Pfeiff., C. Walker & Bloss), which represents the family Diversisporaceae C. Walker & A. Schüßler in the order Diversisporales C. Walker & A. Schüßler (Walker and Schüßler 2004). Consequently, Schüßler and Walker (2010) erected a new genus, Redeckera C. Walker & A. Schüßler, with three species, including R. fulvum (Berk. & Broome) C. Walker & A. Schüßler (formerly G. fulvum) and R. megalocarpum (D. Redecker) C. Walker & A. Schüßler (formerly G. megalocarpum). Unfortunately, the molecular phylogenetic position remained unknown for many other sporocarpic species within the Glomeromycota, which Schüßler and Walker (2010) retained in their original genera, as species of uncertain position.
Despite the lack of molecular evidence, Oehl et al. (2011) transferred some of the sporocarpic Glomus spp. of Glomus sensu lato to other genera of the Glomeromycota. For example, G. canadense (Thaxt.) Trappe & Gerd. and G. versiforme (P. Karst.) S.M. Berch were accommodated in the genera Redeckera and Diversispora C. Walker & A. Schüßler (Diversisporaceae), respectively, because the morphology of the subtending hyphal wall and the spore wall at the spore base in these species clearly differed from the type species of the genus Glomus, G. macrocarpum, (Schüßler and Walker 2010). These transfers were probably correct, but many glomoid spore-producing species of the Glomeromycota do not have a synapomorphy, i.e., a unique morphological character defining a monophyletic group, or have synapomorphies that are invisible under a compound microscope (Błaszkowski et al. 2018a). Therefore, the taxonomic placement of such species within the Glomeromycota is impossible without molecular phylogenetic analyses of sequences of loci with high species resolution. Such well-tested combination of loci include the partial small subunit (SSU), i.e., 18S, rRNA gene; the nuclear internal transcribed spacer 1 (ITS1), 5.8S rRNA gene, and internal transcribed spacer 2 (ITS2) of the rDNA (together called ITS); and the partial large subunit (LSU), i.e., 28S, rRNA gene (together referred to as SSU-ITS-LSU rDNA), as well as the largest subunit of RNA polymerase II (RPB1) gene, which have been recommended as the basis for AM fungal DNA barcoding (Krüger et al. 2009; Stockinger et al. 2014; de Souza et al. 2018).
The fact that fungi-producing glomoid spores in unorganized sporocarps with a peridium constitute a small proportion of the Glomeromycota species and that they are some of the least-known groups of fungi in the phylum with respect to ecophysiology, trophic status, and molecular phylogeny, is mainly the result of the difficulty in collecting these species and growing them in single-species cultures. The collection of sporocarps usually requires the use of specific methods that are much more time-consuming and taxing than the wet-sieving and decanting method (Gerdemann and Nicolson 1963), commonly used to extract hypogeous AMF spores produced singly and in clusters. Because many sporocarpic species are found at or slightly below the soil surface and on plant residues, and their sporocarps are often large enough to be seen with the naked eye, the most frequently used method for collecting these fungi in the field is raking and searching the upper soil layer and buried plant fragments. Such onerous work has certainly discouraged to continue exploration of sporocarpic Glomeromycota. The effective collection of this group of fungi is also depended on the development of a so-called hypogeous instinct (Gerdemann and Trappe 1974).
Of the described species producing glomoid spores in unorganized sporocarps with a peridium, probably only Funneliformis mosseae (T.H. Nicolson & Gerd.) C. Walker & A. Schüßler (formerly G. mosseae T.H. Nicolson & Gerd.), Glomus arborense McGee, G. macrocarpum, G. pallidum I.R. Hall, G. tenerum P.A. Tandy, and G. warcupii McGee were grown in single-species cultures and proven to belong in AMF (Gerdemann and Trappe 1974; Hall 1977; McGee 1986; Schüßler and Walker 2010; http://fungi.invam.wvu.edu/). Attempts at growing other sporocarpic species in culture, for example R. megalocarpum, failed, and, therefore, their mycorrhizal status remains unknown (McGee and Trappe 2002; Redecker et al. 2007). For this reason, it is believed that some of the sporocarpic Glomeromycota may be facultative AMF or even saprotrophs (Goto et al. 2016), which requires further testing.
The failure in obtaining these fungi in culture has significantly limited or prevented the understanding of their morphology, molecular properties, functional diversity, and distribution. Field-collected sporocarps are often incomplete, with partially or completely decomposed external and internal structures, and their spores are often parasitized or empty, and, when DNA is present, it is often degraded. This makes obtaining sequence data from these species for phylogenetic analyses, environmental ecological surveys, and online databases relying on rDNA markers difficult or impossible (pers. observ.).
We found many fungi-producing glomoid spores in unorganized sporocarps and successfully obtained sequence data from them. Detailed morphological and molecular phylogenetic analyses revealed that one of the fungi, collected in Brazil, is an undescribed species of a new genus in the family Glomeraceae, and another fungus, found in Poland, is a new species of the genus Diversispora in the Diversisporaceae. Both fungi are described here.
Materials and methods
Origin of study material, establishment and growth of single-species cultures, extraction of spores, and staining of mycorrhizal structures
Sporocarps of the Brazilian fungus were collected by K. Jobim at two sites of the Adolpho Ducke Forest Reserve (ADFR) in November 2017. The area of the ADFR is ca. 100 km2 and it is located northeast of the city of Manaus (02° 55′ S, 59° 59′ W; Fig. 5). The vegetation of the terra firme forests of the ADFR, which is in the neotropical Campinarana ecoregion, is mainly composed of plant species in the families Arecaceae Bercht. & J.Presl, Burseraceae Kunth, Fabaceae Lindl., Lecythidaceae A. Rich., Malpiguiaceae Juss., and Polygonaceae Juss. (Ribero et al. 1999). Its soils are typically acidic and very poor in nutrients, and are predominantly clayey in the higher sites, becoming sandy as the elevation decreases, as characteristic of podzolic and hydromorphic soils (Chauvel 1982). According to Koppen’s classifications (Peel et al. 2007), the region has a tropical humid climate (type Afi), with an average annual temperature of 26 °C and an annual average rainfall between 1500 and 2500 mm (Alencar et al. 1979; Ribeiro and Adis 1984). Plant species present at the collection sites included, among others, Ecclinusa guianensis Eyma, Eschweilera atropetiolata S.A. Mori, Es.coriaceae (DC.) S. A. Mori, Es. pseudodecolorans S.A. Mori, Es. truncataA.C.Sm., Es. wachenheimii (Benoist) Sandwith, Oenocarpus bacaba Mart., Pouteria anomala (Pires) T.D. Penn., Protium hebetatum D. C. Daly, Pr. paniculatum Engl. and Scleronema micranthum Ducke.
A sporocarp of the Polish fungus was found by Piotr Chachuła in the Pieniny National Park located in southern Poland, on October 10, 2017. The habitat at the collection site (49° 24′ 56.6′ ′N, 20° 19′ 56.5′′ E; 552 m above sea level) was a pine and spruce forest with hazel that had typical soil classified as appropriate typical rendzina (Pancer-Koteja et al. 2004; Zaleski et al. 2016). The dominant herbaceous plant species was Maianthemum bifolium (L.) F. W. Schmidt. Average annual rainfall and temperature in the region where the Pieniny National Park is located are 700–900 mm and 6–8 °C, respectively.
Attempts at establishing and growing single-species cultures were performed using sporocarp fragments, (each containing ca. 20–50 spores) as described by Błaszkowski et al. (2012), using Plantago lanceolata L. as the host plant. Because all attempts at establishing single-species cultures failed, morphological and molecular analyses were performed on spores that were extracted from field-collected sporocarps, using a preparation needle under a stereomicroscope.
Microscopy and nomenclature
At least 50–100 spores of each species were mounted in water, lactic acid, polyvinyl alcohol/lactic acid/glycerol (PVLG, Omar et al. 1979), and a mixture of PVLG and Melzer’s reagent (1:1, v/v) and examined to determine the morphological features and the phenotypic and histochemical characters of their spore wall layers. The preparation of spores for study and photography were as those described previously (Błaszkowski 2012; Błaszkowski et al. 2012). Types of spore wall layers were as defined by Błaszkowski (2012), and Walker (1983). Color names are from Kornerup and Wanscher (1983). Nomenclature of fungi and the authors of fungal names are from the Index Fungorum website (http://www.indexfungorum.org/AuthorsOfFungalNames.htm), except for those referring to the genus represented by a species originally described as Glomus intraradices N.C. Schenck & G.S. Sm. We accepted the arguments presented by Sieverding et al. (2014) and used the generic name Rhizoglomus Sieverd., G.A. Silva & Oehl instead of Rhizophagus P.A. Dang. for the clade with Rh. intraradices (N.C. Schenck & G.S. Sm.) Sieverd., G.A. Silva & Oehl.
Goto and Maia (2006) coined the term “glomerospores” to refer to the spores produced by members of the Glomeromycota because of their unique ontogeny and morphology compared to spores of other phyla of the Kingdom Fungi. In addition, the term unambiguously indicates the taxonomic affiliation of these spores, as do, for example, the ascospores (sexual spores) and ascocarps (fruiting bodies) of the phylum Ascomycota. Consequently, we propose to use the term “glomerocarps” (singular = glomercarp) for conglomerations of glomerospores, previously referred to as sporocarps. We use the terms “glomerocarp(s)” and “glomerospore(s)” only at the beginning of some sections of the paper. In other parts of the text, to avoid unnecessarily increasing their usage, we use the terms “sporocarps,” “sporocarpic,” and “spore(s)”.
Voucher specimens of the new species consisting of dried fragments of glomerocarps in vials and slides with spores mounted in PVLG and a mixture of PVLG and Melzer’s reagent (1:1, v/v) were deposited at the Herbarium of the Federal University of Rio Grande do Norte (UFRN Herbarium; holotype of Sclerocarpum amazonicum), Herbarium of the Instituto Nacional de Pesquisas da Amazônia (INPA Herbarium), Herbarium of the Mycology Department of the Federal University of Pernambuco (URM; isotypes of S. amazonicum), ETH in Zurich, Switzerland (Z+ZT; holotype of Diversispora sporocarpia), and the Department of Ecology, Protection and Shaping of Environment (DEPSE), at West Pomeranian University of Technology, Szczecin (isotypes of both fungi).
Molecular phylogeny, DNA extraction, polymerase chain reaction, cloning, and DNA sequencing
Crude DNA was extracted from eight single spores of each fungus. Details of the treatment of the spores prior to polymerase chain reactions (PCRs), the conditions, and primers used in the PCRs to obtain SSU-ITS-LSU sequences were as those described in Błaszkowski et al. (2015a, 2015b), Krüger et al. (2009), and Symanczik et al. (2014).
In order to obtain RPB1 sequences of the two fungi, nested PCRs were performed in conditions recommended by and with primers designed by Stockinger et al. (2014). The first PCR with DNA of the Brazilian fungus was performed with the primers HS375mix and RPB1-DR1210r, and the second with DR160fmix and RPB1-DR1210r. The first PCR with DNA of the Polish fungus was performed with the primers DR160fmix and HS2680GPr, and the second with HS189GPf and RPB1-DR1210r. For both fungi, in the second nested PCR, 0.5 μl of product from the first amplification was used as template. Cloning and sequencing of PCR products to obtain both types of sequences were performed as described by Błaszkowski et al. (2015a). The sequences were deposited in GenBank (MK036773–MK036789).
Sequence alignment and phylogenetic analyses
Preliminary phylogenetic analyses of sequences of the SSU-ITS-LSU nrDNA region indicated that the Brazilian fungus belonged in the family Glomeraceae, but it was strongly divergent from other representatives of the family. The Polish fungus was closely related to, but distinct from Diversispora epigaea (B.A. Daniels & Trappe) C. Walker & A. Schüßler of the family Diversisporaceae. Consequently, two sequence sets each were assembled, separately for each fungus. One set comprised SSU-ITS-LSU sequences and the second consisted of sequences of the RPB1 gene. Identity values of sequences of the Brazilian fungus only and of the Polish fungus only, as well as the magnitude of sequence divergence of the two fungi from their closest relatives were calculated in BioEdit (Hall 1999). All comparisons were performed on sequences of the same length.
The SSU-ITS-LSU alignment for the Brazilian fungus included sequences of species representing all recognized genera of the Glomeraceae (Redecker et al. 2013; Błaszkowski et al. 2018a, 2018b). The outgroup taxon was Claroideoglomus claroideum (N.C. Schenck & G.S. Sm.) C. Walker & A. Schüßler of the family Entrophosporaceae Oehl & Sieverd. emend. Oehl, Sieverd. Palez. & G.A. Silva. The set did not contain any sequence of the monospecific genera Simiglomus Sieverd., G.A. Silva & Oehl and Viscospora Sieverd., G.A. Silva & Oehl, both erected by Oehl et al. (2011), because (i) their taxonomic status is uncertain; (ii) there is no SSU-ITS-LSU sequence of S. hoi (S.M. Berch & Trappe) G.A. Silva, Oehl & Sieverd. (formerly Glomus hoi S.M. Berch & Trappe) available in public databases; and (iii) both S. hoi and V. viscosa (T.H. Nicolson) Sieverd., Oehl & G.A. Silva (originally described as G. viscosum T.H. Nicolson) differ fundamentally in morphology from the Brazilian fungus (Berch and Trappe 1985; Walker et al. 1995; Redecker et al. 2013; https://invam.wvu.edu/home). The alignment included 112 sequences that represented 36 species of known natural phylogeny, including G. bareae Błaszk. et al., Halonatospora pansihalos (S.M. Berch & Koske) Błaszk. et al. (Błaszkowski et al. 2018b), C. claroideum, and the undescribed Brazilian fungus (Fig. 1). Excluding the Brazilian fungus and C. claroideum, previous molecular phylogenetic analyses placed the other species in eight genera of the Glomeraceae (Redecker et al. 2013; Błaszkowski et al. 2018a, 2018b). Except for G. cf. aggregatum N.C. Schenck & G.S. Sm. and Sclerocystis sinuosa Gerd. & B.K. Bakshi, which were both represented by single LSU sequences, the other species were each characterized by 2–9 SSU-ITS-LSU sequences covering the whole nrDNA segment amplified by the primers of Krüger et al. (2009).
A 50% majority rule consensus phylogram produced from a Bayesian Inference analysis of SSU-ITS-LSU nrDNA sequences of Sclerocarpum amazonicum and 33 other species of AMF in the Glomeraceae, with Claroideoglomus claroideum (Entrophosporaceae) as outgroup. Bayesian Inference posterior probability values ≥ 0.50 and ML bootstrap values ≥ 50% are shown near the branches, respectively. Bar indicates 0.1 expected changes per site per branch
The RPB1 alignment for the Brazilian fungus consisted of RPB1 sequences from species in all genera represented in the SSU-ITS-LSU set, including the genus Claroideoglomus as outgroup (Figs. 1, 2). The alignment comprised 85 sequences, which characterized 32 species of the Glomeraceae, the undescribed Brazilian fungus, and two Claroideoglomus spp. (Fig. 2). In contrast to the SSU-ITS-LSU alignment, the RPB1 alignment did not contain several species for which RPB1 sequences were unavailable, including Dominikia bernensis Oehl et al., Do. compressa (Sieverd. et al.) Oehl et al., D. minuta (Błaszk., Tadych & Madej) Błaszk., Chwat & Kovács, G. cf. aggregatum, Rhizoglomus fasciculatum (Thaxt.) Sieverd., G.A. Silva & Oehl, and Septoglomus jasnowskae Błaszk., Chwat & Ryszka. Except for Sc. sinuosa, Se. deserticola (Trappe, Bloss & J.A. Menge) G.A. Silva, Oehl & Sieverd., Se. viscosum (T.H. Nicolson) C. Walker et al., Rh. intraradices, and the Brazilian fungus, which were each represented by a single RPB1 sequence, each AMF species was characterized by 2–5 sequences.
A 50% majority rule consensus phylogram produced from a Bayesian Inference analysis of RPB1 sequences of Sclerocarpum amazonicum and 30 other species of AMF in the Glomeraceae, with two Claroideoglomus spp. (Entrophosporaceae) as outgroup. Bayesian Inference posterior probability values ≥ 0.50 and ML bootstrap values ≥ 50% are shown near the branches, respectively. Bar indicates 0.05 expected changes per site per branch
The SSU-ITS-LSU alignment for the Polish fungus, which was putatively placed in the family Diversisporaceae based on preliminary analyses, consisted of 86 sequences of 19 species (including the Polish species) in four genera of the Diversisporaceae (Fig. 2). Because the preliminary analyses placed the Polish fungus in the genus Diversispora and indicated that the fungus is most closely related to D. epigaea, the SSU-ITS-LSU alignment included all available sequences of D. epigaea (24), five sequences of the Polish fungus, and 2–3 sequences each of the 14 other Diversispora spp. of known natural phylogeny (Symanczik et al. 2018). The outgroup was two sequences of Pacispora scintillans (S.L. Rose & Trappe) Sieverd. & Oehl ex C. Walker, Vestberg & A. Schüßler of the family Pacisporaceae C. Walker et al. Except for the AM418552 SSU-ITS sequence of Redeckera megalocarpa (D. Redecker) C. Walker & A. Schüßler, the other sequences of the analyzed fungi comprised the entire SSU-ITS-LSU nrDNA region.
The RPB1 alignment for the Polish fungus included 33 sequences of 14 species, including the Polish species, representing four genera of the Diversisporaceae, as well as Sacculospora baltica (Błaszk., Madej & Tadych) Oehl et al. of the family Sacculosporaceae Oehl et al., which served as an outgroup (Fig. 4). Except for D. epigaea and the Polish species, which each had a single RPB1 sequence available in public databases, all other species were represented by two to four RPB1 sequences.
All four sequence alignments were separately aligned with MAFFT v. 7 using the auto option (http://mafft.cbrc.jp/alignment/server/). For the SSU-ITS-LSU alignment, indels were coded by the simple indel coding algorithm (Simmons et al. 2001) as implemented in GapCoder (Young and Healy 2003) and this binary character set was added to the nucleotide alignment, as described and justified in Błaszkowski et al. (2014). The RPB1 set comprised only sequences of the RPB1 gene. Bayesian inference (BI) phylogenetic analyses of the SSU-ITS-LSU and RPB1 alignments were conducted with MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001). In both analyses, the GTR nucleotide substitution model was used, as selected by jModelTest (Posada 2008), using the Akaike information criterion. Four Markov chains were run for 5,000,000 generations, sampling every 100 steps, with a burn-in at 7500 sampled trees. Maximum likelihood (ML) phylogenetic analyses of the alignments were carried out with the raxmlGUI (Silvestro and Michalak 2012) implementation of RAxML (Stamatakis 2014) using the GTRGAMMA algorithm. Then, 1000-replicate rapid bootstrap analysis was performed to determine the support of branches. In both BI and ML analyses of the SSU-ITS-LSU sequences, the analyzed set (the nucleotide alignment plus the binary (indel) character set) was divided into four partitions, knowing that analyses of partitioned data generally increase the accuracy of phylogenetic reconstruction (Lanfear et al. 2012; Nagy et al. 2012). Generated phylogenetic trees were visualized and edited in MEGA6 (Tamura et al. 2013).
Results
General data and phylogeny
The molecular phylogeny for each of the two sporocarpic fungi discussed here was reconstructed on the basis of analyses of two sequence sets, one with sequences of the SSU-ITS-LSU nrDNA region and the second with sequences of the RPB1 gene (Figs. 1–4). The SSU-ITS-LSU alignment of the Glomeraceae for the Brazilian fungus comprised 2348 characters, of which 972 and 892 were variable and parsimony-informative, respectively, as analyzes of the alignment with MEGA6 (Tamura et al. 2013) indicated. The identity values of the four SSU-ITS-LSU sequences of the new fungus ranged from 96.0 to 98.5%. The RPB1 alignment for the Brazilian fungus comprised 2899 characters, of which 1377 and 1288 were variable and parsimony-informative, respectively.
The second SSU-ITS-LSU alignment of the Diversisporaceae for the Polish fungus comprised 1959 characters, of which 710 and 627 were variable and parsimony-informative, respectively. The identity values of the five SSU-ITS-LSU sequences of the fungus ranged from 97.2 to 98.9%. The RPB1 alignment for the Polish fungus comprised 811 characters, of which 223 and 212 were variable and parsimony-informative, respectively.
Bayesian and ML analyses of both the SSU-ITS-LSU and RPB1 alignments placed the Brazilian fungus in a separate clade at the rank of genus that was sister to a clade representing the genus Glomus sensu stricto (Figs. 1, 2). The new generic clade had full support in all analyses (BI = 1, ML = 100%). The common node of the new generic clade and Glomus and the crown node of Glomus itself were also fully or strongly supported.
The identity values of SSU-ITS-LSU sequences of the Brazilian fungus with respect to those of its closest relatives, i.e., G. macrocarpum, G. cf. aggregatum, G. bareae, and G. tetrastratosum (Fig. 1), were 77.1–77.9%, 81.8–82.3%, 76.4–77.4%, and 76.8–77.6%, respectively. The identity values of the RPB1 sequence of the same fungus versus those of G. macrocarpum, G. bareae, and G. tetrastratosum were 86.6%, 85.0%, and 85.2–85.3%, respectively. The molecular identity between the Brazilian fungus and G. cf. aggregatum was not calculated because there is no RPB1 sequence for G. cf. aggregatum in public databases.
Bayesian and ML analyses of both the SSU-ITS-LSU and RPB1 alignments for the Polish fungus placed it as a new species, most closely related to D. epigaea (Figs. 3, 4), thus confirming results of the preliminary analyses mentioned above. In all analyses of both alignments, the clade comprising sequences of the Polish fungus and the common node for that clade and D. epigaea had full or strong support. The identity values of SSU-ITS-LSU and RPB1 sequences of the undescribed fungus compared with SSU-ITS-LSU and RPB1 sequences of D. epigaea were 90.1–96.3% and 98.5%, respectively.
A 50% majority rule consensus phylogram produced from a Bayesian Inference analysis of SSU-ITS-LSU nrDNA sequences of Diversispora sporocarpia and 18 other species of four AMF genera in the family Diversisporaceae, with Pacispora scintillans (Pacisporaceae) as outgroup. Three sequences of D. epigaea are associated with the name Glomus versiforme according to the nomenclature used in GenBank. Bayesian Inference posterior probability values ≥ 0.50 and ML bootstrap values ≥ 50% are shown near the branches, respectively. Bar indicates 0.1 expected changes per site per branch
A maximum likelihood (ML) phylogram produced from analysis of RPB1 sequences of Diversispora sporocarpia, 13 other species of four AMF genera in the family Diversisporaceae, with Sacculospora baltica (Sacculosporaceae) as outgroup. ML bootstrap values ≥ 50% and Bayesian Inference posterior probability values ≥ 0.50 are shown near the branches, respectively. Bar indicates 0.05 expected changes per site per branch
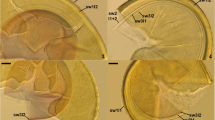
5 Map showing the location of the Adolpho Ducke Forest Reserve (ADFR) and the approximate positions of the two sites in which glomerocarps (= sporocarps) of Sclerocarpum amazonicum were collected; map from http://ppbio.inpa.gov.br/repositorio/dados. 6 Vegetation of site 1 with dense forest and high humidity. 7 Fragments of sporocarps. 8 Sporocarpic spores (sp). Scale bar = 0.5 mm (7). Scale bar = 50 μm (8)
Considering the unambiguously convincing results of the molecular phylogenetic analyses and the comparisons of sequence identities discussed above, we conclude that the Brazilian fungus is a new species representing a new genus in the family Glomeraceae of the Glomeromycota, and the Polish fungus is a new species in the Diversisporaceae. The fungi are described below as Sclerocarpum amazonicum gen. et sp. nov. and D. sporocarpia sp. nov.
Taxonomy
Erection of a new genus
Sclerocarpum B.T. Goto, Błaszk., Niezgoda, Kozłowska & Jobim, gen. nov.
MycoBank No. MB 828316.
Etymology Latin, Sclerocarpum, Sclero (= hard) and carpum (= fruitbody), referring to the very hard glomerocarps (= sporocarps) formed by the type species of the new genus.
Type species:Sclerocarpum amazonicum Jobim, Błaszk., Niezgoda, Kozłowska & B.T. Goto.
Diagnosis Differs from other genera in the Glomeraceae in producing pale-colored, scleroid glomerocarps with hyaline, glomoid spores that have a relatively thick, compared to the small spores, structural laminate spore wall layer, and in having the specific sequences of the nrDNA SSU gene: GGTCTTTGGTTGGTGAGAAG, regions of ITS1: AATGAAATTACGATCATTTA, ITS2: AAAAGATCGATTTTGTCGCCTTTC, AGCTCATCTTTTGAACCTTTC, and LSU gene: TAGCGATACTCGGGTTCTTTGGGCGTACTTTCTCGCT, GCCGAAGTGTTATAGCCTC, CGTAACGGACGGGATC, as well as the RPB1 gene: TTTCATAAGTCATGTAAGCGTTCATC, TAAGCTTACGGAACTTTCTTGATATAG, ACAACCACAAGCATTG, GGTACATCCCGGAGCC, and CAGGGATACTGGAGAG.
Specimens examined: See holotype and isotype specimens of S. amazonicum characterized below.
Genus description: Producing spores in scleroid (very hard to break), epigeous and sub-hypogeous, light- to dark-colored unorganized glomerocarps (= sporocarps) with a peridium and a gleba comprising hyphae and glomoid glomerospores (= spores) with a single subtending hypha (Figs. 7–10, 12). Spores hyaline; globose to subglobose; small, 35–58-μm diam; frequently ovoid (Figs. 8–16). Spore wall composed of two smooth layers (layers 1 and 2), of which the inner layer 2 is laminate and much thicker than the outer layer, forming the spore surface (Figs. 11–16). None of the spore wall layers stains in Melzer’s reagent (Fig. 16). Subtending hypha hyaline, funnel-shaped with a wall consisting of two layers continuous with spore wall layers 1 and 2 (Figs. 10, 11, 14–16). Pore open or occluded by thickening of the subtending hyphal wall, or (rarely) occluded by a straight or slightly invaginated septum continuous with some innermost laminae of spore wall layer 2; septum, positioned at the spore base (Figs. 11, 14–16). Germination directly from the spore wall.
Description of a new species.
Sclerocarpum amazonicum Jobim, Błaszk., Niezgoda, Kozłowska & B.T. Goto, sp. nov.
MycoBank No. MB 828317.
Etymology Latin, amazonicum, referring to the Amazonian ecosystems of South America, in which the species was originally found.
Specimens examined: Brazil, sporocarps found at two sites of the Adolpho Ducke Forest Reserve (02° 55′ S, 59° 59′ W), located in the middle of the Amazonian Forest (Fig. 5), by K. Jobim in November 2017. Holotype. Sporocarps and a slide with spores no. UFRN-FUNGOS 3015, isotypes: a vial with a sporocarp no. 3632, slides with spores no. 3633–3643 (DEPSE), a vial with a sporocarp and slide no. INPA 280748.
Diagnosis: As that of Sclerocarpum.
Description: Glomerospores formed in single epigeous and subhypogeous unorganized glomerocarps. Glomerocarps pale yellow (3A3) to coal (3F1); irregular, (1200–)950 × 1600(− 2000) μm (Figs. 7–9). Peridium thin, yellowish white (3A2) to pale yellow (3A3), only partially covering glomerospore conglomerations (Fig. 9). Gleba pale yellow (3A3) to light yellow (4A4), with hyaline; straight or branched hyphae; 5.0–10.5-μm wide, with a wall 0.8–1.3μm thick; and hundreds of glomerospores (= spores; Figs. 7–10, 12). Spores arise blastically at the tip of sporogenous hyphae (Figs. 10, 11, 14–16). Spores hyaline; globose to subglobose; (35–)48(− 58) µm diam; frequently ovoid; 38–52 × 44–63 μm; with one subtending hypha (Figs. 8–16). Spore wall composed of two layers (Figs. 11–16). Layer 1, forming the spore surface, uniform (not containing visible sublayers), permanent, semi-flexible, smooth, (0.8–)1.3(− 1.5)μm thick, tightly adherent to the upper surface of layer 2 (Figs. 11–16). Layer 2 laminate, permanent, semi-flexible, smooth, (4.8–)6.5(− 9.0)μm thick; consisting of very thin, < 0.5μm thick, laminae, tightly adherent to each other, not separating even in vigorously crushed spores (Figs. 11–16). None of the spore wall layers stains in Melzer’s reagent (Figs. 16). Subtending hypha hyaline; straight or recurved, funnel-shaped; (11.0–)14.9(−19.0)μm wide at the spore base (Figs. 10, 11, 14–16); not braking in crushed spores. Wall of subtending hypha hyaline; (4.3–)7.0(−9.3)μm thick at the spore base; consisting of two layers continuous with spore wall layers 1 and 2 (Figs. 11, 14–16). Pore (1.2–)1.8(− 2.8)μm wide at the spore base, open or occluded by thickening of the subtending hyphal wall, or (rarely) occluded by a straight or slightly invaginated septum continuous with some innermost laminae of spore wall layer 2; septum ca. 1.5–2.3μm thick, positioned at the spore base (Figs. 11, 14–16). Spore content of hyaline oily substance. Germination directly from the spore wall.
Mycorrhizal associations. No molecular analyses were performed on roots of the plant species that grew in the places where the sporocarps of S. amazonicum were found. Attempts to grow S. amazonicum in single-species cultures with P. lanceolata as the host plant failed.
Distribution and habitat. To date, S. amazonicum has been found only in the Adolfo Ducke Forest Reserve located in the middle of the Amazonian Forest (Fig. 5). The geographical position and the composition of vegetation of the sites, in which S. amazonicum was found are characterized in the section “Materials and Methods.” The soil surface of the sites where the epigeous and subhypogeous sporocarps of the fungus occurred was covered with a large amount of organic matter.
BLAST searches indicated that this species has not been recorded in the world before. The highest identities of SSU-ITS-LSU and RPB1 sequences of S. amazonicum compared with sequences of these two loci available in GenBank were only 90% and 88%, respectively.
Diversispora sporocarpia Chachuła, Mleczko, Zubek, Niezgoda, Kozłowska, Jobim, B.T. Goto & Błaszk. sp. nov.
MycoBank No. MB 828318.
Etymology Latin, sporocarpia, referring to the sporocarp formed by the species.
Specimens examined: Poland, sporocarp found in the Pieniny National Park (49° 24′ 56.6′ ′N, 20° 19′ 56.5′ ′E) by Piotr Chachuła on October 10, 2017. Holotype. A slide with spores no. ZT Myc 59592 (Z+ZT), isotypes: a vial with a sporocarp no. 3644 and slides with spores no. 3645–3649 (DEPSE).
Diagnosis: Differs from other Diversispora spp. in producing compact unorganized glomerocarps consisting of a peridium and a gleba comprising spores and hyphae, in the phenotypic characters of spore wall layers, and in the nucleotide composition of sequences of the SSU-ITS-LSU nrDNA region and the RPB1 gene.
Description: Glomerospores formed in a compact epigeous unorganized glomerocarp consisting of a peridium and a gleba. Glomerocarp grayish orange (6B3) to light brown(6D8); ovate; 6 × 4 mm (Fig. 17). Peridium thin, hyaline to light yellow (4A4), only partially covering spore conglomerations (Fig. 17). Gleba yellow (3A6) to light brown (5D8), with hyaline, straight or branched hyphae; 12.5–22.3μm wide, with a wall 1.0–1.5μm thick; and hundreds of spores (Figs. 17, 18). Spores arise blastically at the tip of sporogenous hyphae (Figs. 23, 24). Spores yellow (3A6) to light brown (5D8); usually ovoid; 110–140 × 140–190 μm; rarely globose to subglobose; (120–)136(−148)μm diam; with one subtending hypha (Figs. 17, 23, 24). Spore wall composed of four layers (Figs. 19, 20, 21, 22, 23, 24). Layer 1, forming the spore surface, semi-permanent, semi-flexible, smooth when intact, more or less roughened when deteriorated, rarely completely sloughed off, pale yellow (4A3), (0.8–)1.4(−2.3)μm thick when intact (Figs. 19–24). Layer 2 uniform (not divided into visible sublayers), permanent, semi-flexible, yellow (3A6) to light brown (5D8), (1.0–)1.5(−2.0)μm thick (Figs. 19–24). Layers 1 and 2 tightly adherent to and not separating from each other even in vigorously crushed spores (Figs. 19–24). Layer 3 laminate, permanent, semi-flexible, hyaline to yellowish white (4A2), (5.0–)7.6(−9.8)μm thick, consisting of very thin, < 0.5μm thick, laminae, frequently separating from each other in vigorously crushed spores; when colored, the closer to the lower surface, the sublayers of layer 3 lighten and become colorless (Figs. 19–24); layer 3 occasionally separates from the lower surface of layer 2 in vigorously crushed spores (Fig. 22). Layer 4 uniform, permanent, flexible, hyaline, 0.4–0.7μm thick, loosely associated with the lower surface of layer 3, clearly visible in slightly crushed spores, where it usually is separated from the intact laminate layer 3 (Figs. 19–24). Layers 1–4 are not staining in Melzer’s reagent (Figs. 23, 24). Subtending hypha pastel yellow (3A4) to grayish yellow (3B8); straight or recurved, cylindrical, rarely slightly funnel-shaped; (10.2–)12.2(− 15.7)μm wide at the spore base (Figs. 23, 24); not braking in crushed spores. Wall of subtending hypha pastel yellow (3A4) to grayish yellow (3B8); (3.8–)4.9(− 5.5)μm thick at the spore base; consisting of four layers continuous with spore wall layers 1–4; subtending hyphal wall layer (shwl) 1 usually highly deteriorated or completely sloughed off; shwl 3 extending 6.4–16.0 μm below the spore base; shwl4 usually difficult to see (Figs. 23, 24). Pore (0.8–)3.0(− 5.0)μm wide at the spore base, occluded (i) by thickening of the subtending hyphal wall, (ii) by a straight or slightly invaginated septum connecting the inner surfaces of subtending hyphal wall layer 4, (iii) by both the structures, or (iv) (rarely) by a septum continuous with the innermost laminae of spore wall layer 3, rarely open; septum 1.3–5.5μm thick, positioned at or up to 7.5 μm below the spore base (Figs. 23, 24). Spore content of hyaline oily substance. Germination unknown.
Mycorrhizal associations. No molecular analyses were performed on roots of M. bifolium that grew in the area where the sporocarp of D. sporocarpia was found. Attempts to grow D. sporocarpia in single-species cultures with P. lanceolata as the host plant failed.
Distribution and habitat. The forest of the Pieniny National Park (pine and spruce forest with hazel), whose geographical position and climate are characterized in section “Materials and Methods,” is the sole reported location of D. sporocarpia. However, a BLAST search of the D. sporocarpia SSU-ITS-LSU sequences yielded two similar sequences with 98% identity (HE775321, HE775336), which were detected in environmental sequencing conducted in the Czech Republic (Kohout et al. 2014) and may represent the same species. There are no RPB1 sequences in GenBank that would suggest conspecificity with D. sporocarpia.
Discussion
The validity of the erection of the new monospecific genus Sclerocarpum in the family Glomeraceae unambiguously proved the results discussed above. First, phylogenetic analyses placed S. amazonicum in a clade located equivalently to other clades designated as genera in the Glomeraceae (Figs. 1, 2). Second, the divergences of the SSU-ITS-LSU sequences of S. amazonicum from the SSU-ITS-LSU sequences of G. bareae, G. macrocarpum, and G. tetrastratosum, which grouped in a sister clade in the reconstructed phylogenetic tree (Fig. 1), and the divergence of the RPB1 sequence of S. amazonicum from RPB1 sequences of the same three Glomus spp., which also positioned in a sister clade (Fig. 2), were 22.1–23.6% and 14.4–15.0%, respectively. Thus, the divergences were much or clearly larger than divergences of sequences of the same loci of other phylogenetically characterized neighbors belonging to different genera of the Glomeraceae (Błaszkowski et al. 2018a, 2018b). For example, in the Glomeraceae SSU-ITS-LSU phylogenetic tree (Fig. 1), the neighboring Rhizoglomus irregulare FM865555 and Sclerocystis sinuosa FJ461846 sequences differed by 10.5%, and the difference between the neighboring G. tetrastratosum KM056665 and S. viscosum HF548854 sequences was 20.0%. In the RPB1 tree (Fig. 2), the neighboring Sclerocystis pubescens HG316023 and Rh. irregulare HG315983 sequences differed by 14.4%.
Sclerocarpum amazonicum. 9 Glomerocarps (= sporocarp) with glomerospores (= spores; sp) and peridium (p). 10 Glebal spores (sp) with subtending hyphae (sh) and glebal hyphae (gh). 11 Subtending hyphal wall layers (shwl) 1 and 2 continuous with spore wall layers (swl) 1 and 2 and glebal hyphae (gh). 12, 13 Spore wall layers (swl) 1 and 2 and glebal hyphae (gh). 14–16 Subtending hyphal wall layers (shwl) 1 and 2 continuous with spore wall layers 1 and 2; note the pore of the subtending hypha is closed by thickened shwl2 (14, 15), and it is closed by a straight septum continuous with the innermost laminae of swl2 (16). 9 Dry herbarium specimen. 10–15 Spores and glebal hyphae in PVLG. 16 Spore in PVLG + Melzer’s reagent. 9 Light microscopy. 10–16 Differential interference microscopy. Scale bars = 100 μm (9), 20 μm (10), 10 μm (11–16)
Diversispora sporocarpia. 17 Glomerocarp (= sporocarp) with glomerospores (= spores; sp) and peridium (p). 18 Glebal hyphae (gh). 19–22 Spore wall layers (swl) 1–4. 23, 24. Subtending hyphal wall layers (shwl) 1–4 continuous with spore wall layers (swl) 1–4; note the pore of the subtending hypha is closed by thickened shwl3 (23) or it is closed by a straight septum (s) connecting the inner surfaces of shwl4 (24). 17 Dry herbarium specimen. 18–22 Spores in PVLG. 23, 24 Spores in PVLG+Melzer’s reagent. 17 Light microscopy. 18–24. Differential interference microscopy. Scale bars = 200 μm (17), 10 μm (18–22)
The morphological features that clearly distinguish S. amazonicum, the so far sole known species of the genus Sclerocarpum, from representatives of other genera of the Glomeromycota that produce glomoid spores in unorganized sporocarps with a peridium are the small, hyaline, thick-walled spores and the cylindrical or funnel-shaped spore subtending hypha, whose wall layers clearly arise below the spore base and are continuous with spore wall layers (Figs. 7–16).
Surprisingly, our molecular phylogenetic analyses, which also considered many AMF species that produce hyaline, small glomoid spores, indicated that the most closely related species to S. amazonicum are G. cf. aggregatum, G. bareae, G. macrocarpum, and G. tetrastratosum (Figs. 1, 2). Of these, only G. macrocarpum is known to form spores in unorganized sporocarps with a peridium, although spores of the species may also be produced singly (McGee and Trappe 2002; Schüßler and Walker 2010; Błaszkowski 2012). Most importantly, spores of the latter four species are colored [pastel yellow (2A4) to yellowish brown (5D8)], are much larger (up to 4.1-fold when globose), and their spore wall consist of three to four layers, with the exception of the spore wall of G. macrocarpum, which is also two-layered (Berch and Fortin 1983; Błaszkowski 2012). However, spore wall layer 1 of G. macrocarpum, which forms the spore surface, similarly as spore wall layer 1 of the other three Glomus spp., is a short-lived structure that is usually mostly or completely sloughed off in mature spores, and it stains pink (11A5) to deep Magenta (14D8) in Melzer’s reagent. Spore wall layer 1 of S. amazonicum is presumably permanent, as it was present in all specimens examined by us, and it is nonreactive in Melzer’s (Figs. 11–16).
Additional species that probably belong to the genus Sclerocarpum are G. pallidum I.R. Hall, G. segmentatum Trappe, Spooner & Ivory, and G. tenerum P.A. Tandy. All three produce unorganized sporocarps with a peridium and glomoid spores. Unfortunately, the natural phylogenies of these species remain unknown and, therefore, the determination of their molecular relationship to S. amazonicum is impossible. However, other morphological characters prove that S. amazonicum differs fundamentally from the three Glomus spp. Spores of G. pallidum are colored [orange (4A3) to pale orange (5A3)], their two-layered spore wall is 1.6–2.3-fold thinner, and their spore wall layer 1 is evanescent, and frequently completely sloughed off in mature spores (Błaszkowski 2012). Moreover, the subtending hypha of G. pallidum spores is more uniform in shape (cylindrical to slightly flared) and, at the spore base, it is 1.6–2.2-fold narrower, and has a 2.9–6.1-fold thinner wall. Spores of G. segmentatum are 1.6–1.9-fold larger when globose and their spore wall consists of three layers, of which layer 1, covering the upper surface of the structural laminate layer 2, is frequently completely sloughed off in mature specimens (Oehl et al. 2011; Błaszkowski 2012). In addition, the laminate spore wall layer 2 of G. segmentatum occasionally stains reddish white (9A2) in Melzer’s reagent (Błaszkowski 2012). Most importantly, unique to G. segmentatum is the formation of sporocarps composed of tough segments, of which each segment, containing crowded spores, is enclosed with a peridium (Trappe 1979; Furrazola et al. 2016). Spores of G. tenerum are much darker (yellow orange to brown) and 1.3–3.4-fold larger when globose (McGee and Trappe 2002).
Our molecular phylogenetic analyses showed that the closest relative of D. sporocarpia is D. epigaea (Figs. 3, 4). The novelty of D. sporocarpia was also confirmed by the large divergences of its SSU–ITS–LSU and RPB1 sequences when compared to D. epigaea, which were 3.7–9.9% and 1.5%, respectively.
Morphologically, D. sporocarpia also differs clearly from D. epigaea. The main distinctions reside in the number and phenotypic features of their spore wall components. The spore wall of D. sporocarpia consists of four layers (Figs. 19–24), whereas the D. epigaea spore wall is three-layered (Schüßler et al. 2011). Both species share the presence of a penultimate structural laminate layer and a thin, flexible innermost layer in the spore wall. However, the structural laminate layer 3 of D. sporocarpia is surrounded by a semi-permanent, 0.8–2.3μm thick layer 1, forming the spore surface, and by a uniform, permanent, 1.0–2.0μm thick layer 2 that directly covers the upper surface of spore wall layer 3 (Figs. 19–24). In the D. epigaea spore wall, the structural laminate layer is the second layer of this wall and it is covered with an evanescent outermost layer 1, which is up to 1μm thick when intact (Schüßler et al. 2011). In addition, the subtending hypha of D. sporocarpia spores may be up to 1.6-fold wider at the spore base.
As we mentioned in the section “Introduction,” species of AMF that produce glomoid spores in unorganized sporocarps with a peridium are one of the least-known fungal groups of the Glomeromycota. Such sporocarpic species apparently have much higher ecological (e.g., physical and biological soil properties, the composition of plant communities, temperature, humidity) and temporal requirements for fruiting than other groups of the Glomeromycota. Therefore, for herbaria and institutions with fungal collections that are interested in increasing the representation of these members of the Glomeromycota, the only solution is to pursue the collection of their fresh specimens, particularly in regions that are probably more conductive to the development of these sporocarpic species. According to Hawksworth and Rossman (1997), Hawksworth (2001), and Blackwell (2011), tropical forests are such regions, and the exploration of tropical forests have resulted in the discovery of many sporocarpic fungi (Sulzbacher et al. 2016, 2017). These include the recently newly described species of the Glomeromycota from Brazil and Cuba (Torre-Arias et al. 2017), AMF species found in these countries, but previously rarely recorded in the world (Goto and Maia 2005), as well as the new Brazilian fungus, S. amazonicum, characterized in this paper.
However, based on literature data and our own observations, we conclude that the Glomeromycota fungi that produce glomoid spores in unorganized sporocarps with a peridium probably have a worldwide distribution and they likely occur not only in undisturbed high-humidity habitats that are rich in organic matter, but also in highly degraded soils (McGee and Trappe 2002). It is difficult to speculate about the worldwide frequency of sporocarpic Glomeromycota. The relatively infrequent records of these fungi are likely not due to the rarity of their occurrence. Rather, they are the result of rare use of the specific and labor-intensive methods necessary to find their sporocarps. Another reason is the lack of experienced mycologists interested in deepening the knowledge of this group of fungi, which also creates a lot of difficulties in molecular studies (pers. observ.). Finally, these fungi will have little to no practical applications (e.g. as commercial inocula) because they are difficult to grow in culture and there is no certainty that all of them are typical mycorrhizal symbionts.
References
Alencar JC, Almeida RA, Fernandes NP (1979) Fenologia de espécies florestais em floresta tropical úmida de terra firme na Amazônia Central. Acta Amazon 9:163–198. https://doi.org/10.1590/1809-43921979091163
Berch SM, Fortin JA (1983) Lectotypification of Glomus macrocarpum and proposal of new combinations: Glomus australe, Glomus versiforme and Glomus tenebrosum (Endogonaceae). Can J Bot 61:2608–2617. https://doi.org/10.1139/b83-287
Berch SM, Trappe JM (1985) A new species of Endogonaceae, Glomus hoi. Mycologia 77:654–657. https://doi.org/10.2307/3793364
Blackwell M (2011) The fungi: 1, 2, 3 … 5.1 million species? Am J Bot 98(3):426–438. https://doi.org/10.3732/ajb.1000298
Błaszkowski J (2012) Glomeromycota. W. Szafer institute of botany, Polish Academy of Sciences, Kraków
Błaszkowski J, Chwat G, Góralska A, Bobrowska-Chwat A (2015a) Glomus tetrastratosum, a new species of arbuscular mycorrhizal fungi (Glomeromycota). Mycoscience 56:280–286. https://doi.org/10.1016/j.myc.2014.08.003
Błaszkowski J, Chwat G, Góralska A, Ryszka P, Kovács GM (2015b) Two new genera, Dominikia and Kamienskia, and D. disticha sp. nov. in Glomeromycota. Nova Hedwigia 100:225–238. https://doi.org/10.1127/nova_hedwigia/2014/0216
Błaszkowski J, Chwat G, Góralska A, Ryszka P, Orfanoudakis M (2014) Septoglomus jasnowskae and Septoglomus turnauae, two new species of arbuscular mycorrhizal fungi (Glomeromycota). Mycol Prog 13(4):999–1009. https://doi.org/10.1007/s11557-014-0985-z
Błaszkowski J, Kovács GM, Gáspár BK, Balázs TK, Buscot F, Ryszka P (2012) The arbuscular mycorrhizal Paraglomus majewskii sp. nov. represents a new distinct basal lineage in Paraglomeraceae (Glomeromycota). Mycologia 104(1):148–156. https://doi.org/10.3852/10-430
Błaszkowski J, Niezgoda P, Goto BT, Dalpé Y (2018a) A new genus, Oehlia with Oehlia diaphana comb. nov. and an emended description of Rhizoglomus vesiculiferum comb. nov. in the Glomeromycotina. Nova Hedwigia. https://doi.org/10.1127/nova_hedwigia/2018/0488
Błaszkowski J, Niezgoda P, Goto BT, Kozłowska A (2018b) Halonatospora gen. nov. with H. pansihalos comb. nov. and Glomus bareae sp. nov. (Glomeromycota; Glomeraceae). Botany. https://doi.org/10.1139/cjb-2018-0107
Chauvel A (1982) Os latossolos amarelos, álicos, argilosos dentro dos ecossistemas das bacias experimentais do INPA e da região vizinha. Acta Amazo 12:47–60. https://doi.org/10.1590/1809-43921982123S047
de Souza FA, Silva IR, Barros-Barreto MBB, Oehl F, Goto BT, Maia LC (2018) Racocetra crispa (Glomeromycotina) delimited by integrative evidence based on morphology, long continuous nuclear rDNA sequences and phylogeny. Mycol Prog 17:999–1011. https://doi.org/10.1007/s11557-018-1410-9
Furrazola E, Torre-Arias Y, Thoen D, Berbara RLL, Jobim K, Goto BT (2016) Glomus segmentatum, rediscovery of a rare epigeous sporocarpic fungus to Cuba. Cur Res Environ Appl Mycol 6:143–149. https://doi.org/10.5943/cream/6/2/8
Gerdemann JW, Nicolson TH (1963) Spores of mycorrhizal Endogone species extracted from soil by wet-sieving and decanting. Trans Brit Mycol Soc 46:235–244. https://doi.org/10.1016/S0007-1536(63)80079-0
Gerdemann JW, Trappe JM (1974) The Endogonaceae in the Pacific northwest. Mycol Mem 5:1–76
Goto BT, Bezerra JL, Maia LC (2016) Sclerocystis coremioides (Glomeromycota) formando esporocarpos epígeos em substratos orgânicos de cacaueiro na Mata Atlântica da Bahia. Agrotrópica 28:23–28. https://doi.org/10.21757/0103-3816.2016v28n1p23-28
Goto BT, Maia LC (2005) Sporocarpic species of arbuscular mycorrhizal fungi (Glomeromycota), with a new report from Brazil. Acta Bot Bras 19(3):633–637. https://doi.org/10.1590/S0102-33062005000300025
Goto BT, Maia LC (2006) Glomerospores: a new denomination for the spore of Glomeromycota, a group molecularly distinct from the Zygomycota. Mycotaxon 96:129–132
Hall IR (1977) Species and mycorrhizal infections of New Zealand Endogonaceae. Trans Br Mycol Soc 68:341–356. https://doi.org/10.1016/S0007-1536(77)80186-1
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hawksworth DL (2001) The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol Res 105(12):1422–1432. https://doi.org/10.1017/S0953756201004725
Hawksworth DL, Rossman AY (1997) Where are all the undescribed fungi? Phytopathology 87(9):888–891. https://doi.org/10.1094/PHYTO.1997.87.9.888
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. https://doi.org/10.1093/bioinformatics/17.8.754
Kohout P, Sudová R, Janoušková M, Čtvrtlíková M, Hejda M, Pánková H, Slavíková R, Stajerová K, Vosátka M, Sýkorová Z (2014) Comparison of commonly used primer sets for evaluating arbuscular mycorrhizal fungal communities: is there a universal solution? Soil Biol Biochem 68:482–493. https://doi.org/10.1016/j.soilbio.2013.08.027
Kornerup A, Wanscher JH (1983) Methuen handbook of colour, 3rd edn. E. Methuen, London
Krüger M, Stockinger H, Krüger C, Schüßler A (2009) DAN-based level detection of Glomeromycota: one PCR primer set for all arbuscular mycorrhizal fungi. New Phytol 183:212–223. https://doi.org/10.1111/j.1469-8137.2009.02835.x
Lanfear R, Calcott B, Ho SYW, Guindon S (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29(6):1695–1701. https://doi.org/10.1093/molbev/mss020
McGee PA (1986) Further sporocarpic species of Glomus (Endogonaceae) from South Australia. Trans Brit Mycol Soc 87:123–129. https://doi.org/10.1016/S0007-1536(86)80011-0
McGee PA, Trappe JM (2002) The Australian zygomycetous mycorrhizal fungi. II. Further Australian sporocarpic Glomaceae. Aust Syst Bot 15:115–124. https://doi.org/10.1071/SB00038
Morton JB (1988) Taxonomy of VA mycorrhizal fungi: classification, nomenclature. and identification Mycotaxon 32:267–324
Morton JB, Benny GL (1990) Revised classification of arbuscular mycorrhizal fungi (Zygomycetes): a new order, Glomales, two new suborders, Glomineae and Gigasporineae, and two new families, Acaulosporaceae and Gigasporaceae, with an emendation of Glomaceae. Mycotaxon 37:471–491
Nagy LG, Kocsube S, Csana Z, Kovacs GM, Petkovits T, Lgyi CV, Papp T (2012) Re-mind the gap! Insertion–deletion data reveal neglected phylogenetic potential of the nuclear ribosomal internal transcribed spacer (ITS) of fungi. PLoS One 7(11):e49794. https://doi.org/10.1371/journal.pone.0049794
Oehl F, da Silva GA, Goto BT, Sieverding E (2011) Glomeromycota: three new genera and glomoid species reorganized. Mycotaxon 116:75–120. https://doi.org/10.5248/116.75
Omar MB, Bollan L, Heather WA (1979) A permanent mounting medium for fungi. Bull Br Mycol Soc 13:31–32. https://doi.org/10.1016/S0007-1528(79)80038-3
Pancer-Koteja E, Bodziarczyk J, Holeksa J, Piątek G, Różański W, Szwagrzyk J, Kaźmierczakowa R, Dubiel E, Perzanowska J, Wróbel I, Vončina G, Zarzycki J (2004) Vegetation map of the Pieniny National Park, 1998–2001
Peel MC, Finlayson BL, McMahon TA (2007) Update world map of the Köppen-Geiger climate classification. Hydrol Earth Syst Sc 11:1633–1644. https://doi.org/10.5194/hess-11-1633-2007
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. https://doi.org/10.1093/molbev/msn083
Redecker D, Raab PA, Oehl F, Camacho FJ, Courtecuisse R (2007) A novel clade of sporocarp-forming species of glomeromycotan fungi in the Diversisporales lineage. Mycol Prog 6:35–44. https://doi.org/10.1007/s11557-007-0524-2
Redecker D, Schüßler A, Stockinger H, Stürmer SL, Morton JB, Walker C (2013) An evidence-based consensus for the classification of arbuscular mycorrhizal fungi (Glomeromycota). Mycorrhiza 23:515–531. https://doi.org/10.1007/s00572-013-0486-y
Ribeiro MNG, Adis J (1984) Local rainfall variability—a potential bias for bioecological studies in the Central Amazon. Acta Amazon 14:159–174. https://doi.org/10.1590/1809-43921984142174
Schüßler A, Schwarzott D, Walker C (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Myc Res 105:1413–1421. https://doi.org/10.1017/S0953756201005196
Schüßler A, Krüger M, Walker C (2011) Revealing natural relationships among arbuscular mycorrhizal fungi: culture line BEG47 represents Diversispora epigaea, not Glomus versiforme. PLoS One 6(8):e23333. https://doi.org/10.1371/journal.pone.0023333
Schüßler A, Walker C (2010) The Glomeromycota: a species list with new families and new genera. Gloucester
Sieverding E, da Silva GA, Berndt R, Oehl F (2014) Rhizoglomus, a new genus of the Glomeraceae. Mycotaxon 129(2):373–386. https://doi.org/10.5248/129.373
Silvestro D, Michalak I (2012) raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 12:335–337. https://doi.org/10.1007/s13127-011-0056-0
Simmons MP, Ochoterena H, Carr TG (2001) Incorporation, relative homoplasy, and effect of gap characters in sequence-based phylogenetic analyses. Syst Biol 50:454–462. https://doi.org/10.1080/106351501300318049
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Stockinger H, Peyret-Guzzon M, Koegel S, Bouffaud M-L, Redecker D (2014) The largest subunit of RNA polymerase II as a new marker gene to study assemblages of arbuscular mycorrhizal fungi in the field. PLoS One 9(10):e107783. https://doi.org/10.1371/journal.pone.0107783
Sulzbacher MA, Grebenc T, Cabral TS, Giachini AJ, Goto BT, Smith ME, Baseia IG (2016) Restingomyces, a new sequestrate genus from the Brazilian Atlantic rainforest that is phylogenetically related to early-diverging taxa in Trappeaceae (Phallales). Mycologia 108:954–966. https://doi.org/10.3852/15-265
Sulzbacher MA, Grebenc T, Giachini AJ, Baseia IG (2017) Sclerotium-forming fungi from soils of the Atlantic rainforest of northeastern Brazil. Plant ecol evol 150:358–362. https://doi.org/10.5091/plecevo.2017.1148
Symanczik S, Al-Yahya’ei MN, Kozłowska A, Ryszka P, Błaszkowski J (2018) A new genus, Desertispora, and a new species, Diversispora sabulosa, in the family Diversisporaceae (order Diversisporales, subphylum Glomeromycotina). Mycol Prog 17:437–449. https://doi.org/10.1007/s11557-017-1369-y
Symanczik S, Błaszkowski J, Chwat G, Boller T, Wiemken A, Al-Yahya'ei MN (2014) Three new species of arbuscular mycorrhizal fungi discovered at one location in a desert of Oman: Diversispora omaniana, Septoglomus nakheelum and Rhizophagus arabicus. Mycologia 106:243–259. https://doi.org/10.3852/106.2.243
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Tedersoo L, Sanchez-Ramirez S, Koljalg U, Bahram M, Doring M, Schigel D, May T, Ryberg M, Abarenkov K (2018) High-level classification of the fungi and a tool for evolutionary ecological analyses. Fungal Divers 90(1):135–159. https://doi.org/10.1007/s13225-018-0401-0
Torre-Arias Y, Furrazola E, Berbara RLL, Jobim K, Lima JLR, Goto BT (2017) Glomus herrerae, a new sporocarpic species of Glomeromycetes from Cuba. Cur Res Environ Appl Mycol 7:55–160. https://doi.org/10.5943/cream/7/3/2
Trappe JM (1979) Glomus segmentatum sp. nov. Trans Brit Mycol Soc 73:362–363. https://doi.org/10.1016/S0007-1536(79)80127-8
Turrini A, Saran M, Giovannetti M, Oehl F (2018) Rhizoglomus venetianum, a new arbuscular mycorrhizal fungal species from a heavy metal-contaminated site, downtown Venice in Italy. Mycol Prog. https://doi.org/10.1007/s11557-018-1437-y
Walker C (1983) Taxonomic concepts in the Endogonaceae: spore wall characteristics in species descriptions. Mycotaxon 18:443–455
Walker C, Giovannetti M, Avio L, Citernesi AS, Nicolson TH (1995) A new fungal species forming arbuscular mycorrhizas: Glomus viscosum. Mycol Res 99:1500–1506. https://doi.org/10.1016/S0953-7562(09)80799-5
Walker C, Schüßler A (2004) Nomenclatural clarifications and new taxa in the Glomeromycota. Mycol Res 108:979–982. https://doi.org/10.1017/S0953756204231173
Young ND, Healy J (2003) GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinformatics 4(6). https://doi.org/10.1186/1471-2105-4-6
Zaleski T, Mazurek R, Gąsiorek M, Wanic T, Zadrożny P, Józefowska A, Kajdas B, Wężyk P, Szostak M, Usień M, Zięba-Kulawik K, Hawryło P (2016) Mapa gleb ekosystemów leśnych Pienińskiego Parku Narodowego. Kraków-Krościenko nad Dunajcem, Archiwum Pienińskiego Parku Narodowego
Acknowledgments
The authors thank Universidade Federal do Rio Grande do Norte for covering the costs of J. Błaszkowski’s stay as visiting professor (Internationalization incentive program PPg/UFRN/2017). Thanks are due to the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for providing PhD scholarships to K. Jobim. The authors also thank Fernando Sarti Andriolli, researcher of Instituto Nacional de Pesquisas da Amazônia, for the assistance provided in the search for sporocarps in the Adolpho Ducke Forest Reserve (ADFR), and Carolina Castilho for the identification of plant species of the ADFR. Fieldwork infrastructure and logistic support in the Amazon were provided by the National Institute for Amazonian Biodiversity (INCT-CENBAM), Brazilian Program for Biodiversity Research (PBBio), the program Large-Scale Biosphere-Atmosphere Experiment on Amazonia (LBA), and Fundação Amparo à Pesquisa do Estado do Amazonas (FAPEAM). The authors are grateful to the Editor, Associate Editor, and two anonymous reviewers for many helpful comments on the previous versions of this paper.
Funding
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), which provided research grants to BT Goto and financial support for the project “Sporocarpic species of arbuscular mycorrhizal fungi (Glomeromycota): taxonomy, systematic and evolution” (proc. 408011-2016-5). The research was also funded, in part, by the Institute of Botany at the Jagiellonian University in Kraków, projects no. K/ZDS/006300, K/ZDS/007340, and K/ZDS/008054.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Marco Thines
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
OpenAccess This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Jobim, K., Błaszkowski, J., Niezgoda, P. et al. New sporocarpic taxa in the phylum Glomeromycota: Sclerocarpum amazonicum gen. et sp. nov. in the family Glomeraceae (Glomerales) and Diversispora sporocarpia sp. nov. in the Diversisporaceae (Diversisporales). Mycol Progress 18, 369–384 (2019). https://doi.org/10.1007/s11557-018-01462-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-018-01462-2