Abstract
Phylogenetic analyses of sequences of the SSU–ITS–LSU nrDNA segment and the RPB1 gene showed that the arbuscular mycorrhizal fungus originally described as Diversispora omaniana does not belong to the genus Diversispora, but represents a separate clade at the rank of genus in the family Diversisporaceae of the order Diversisporales. The closest natural relatives of the fungus proved to be species of the genera Corymbiglomus and Redeckera. Consequently, the new genus was named Desertispora, and Di. omaniana was renamed De. omaniana comb. nov. In addition, the morphological and histochemical features of spores and mycorrhizal structures of a new Diversispora sp., Di. sabulosa, were described and the closest relatives of the species were determined based on phylogenetic analyses of sequences of the two loci mentioned above. The new fungus was grown in single-species cultures established from spores extracted from a trap culture inoculated with a mixture of the rhizosphere soil and root fragments of Ammophila arenaria that had colonized maritime sand dunes of the Curonian Spit located in the north of Lithuania. Diversispora sabulosa was never found before in many different sites of the world which were sampled during the last 34 years by the last author of the paper. Also, the lack of molecular sequences in public databases of identity ≥ 97% to sequences of Di. sabulosa suggests that the fungus is rare on the Earth.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The subphylum Glomeromycotina (C. Walker & A. Schüßler) Spatafora & Stajich (phylum Mucoromycota Doweld; Spatafora et al. 2016), comprising arbuscular mycorrhizal fungi (AMF), consists of four orders, of which the Diversisporales sensu C. Walker & A. Schüßler is distinguished by possessing species most diverse in morphology and histochemical properties of spores and mycorrhizal structures, and molecular phylogeny (Walker and Schüßler 2004; Schüßler and Walker 2010). Oehl et al. (2011a) divided the Diversisporales to erect Gigasporales Sieverd. et al. ord. nov. comprising AMF forming spores from a bulbous sporogenous hypha, called gigasporoid spores. Redecker et al. (2013) rejected the proposal as unsupportable, but Castillo et al. (2016) and Pagano et al. (2016) accepted the erection of the Gigasporales. In the present study, we have not checked the accuracy of the decisions, but tested the hypothesis that the species originally described as Diversispora omaniana Symanczik, Błaszk. & Al-Yahya’ei (Symanczik et al. 2014) represents an undescribed taxon in the Diversisporales. In order to determine the rank and position of the taxon, we concluded that the molecular properties of the species should be compared with representatives of as many families of the Diversisporales as possible. Therefore, we consider here the composition of the Diversisporales according to its original definition (Walker and Schüßler 2004; Schüßler and Walker 2010).
The widely understood Diversisporales contains five families, among which is the Diversisporaceae C. Walker & A. Schüßler with five genera. Three of the genera, Corymbiglomus Błaszk. & Chwat, Diversispora C. Walker & A. Schüßler, and Redeckera, are represented by species forming spores at the tip of a cylindrical or funnel-shaped sporogenous hypha (Schüßler and Walker 2010; Błaszkowski 2012; Błaszkowski and Chwat 2013). Spores of the monospecific genera Otospora Oehl, Palenz. & N. Ferrol and Tricispora Oehl et al. arise laterally on and inside, respectively, the neck of a sporiferous saccule (Palenzuela et al. 2008; Oehl et al. 2011c). Thus, the spore formation modes of O. bareae Palenz., N. Ferrol & Oehl and T. nevadensis (Palenz. et al.) Oehl et al., the latter originally described as Entrophospora nevadensis Palenz. et al. (Palenzuela et al. 2010), resemble those of spores of Acaulospora laevis Gerd. & Trappe and Entrophospora infrequens (I.R. Hall) R.N. Ames & R.W. Schneid., respectively, the type species of the genera Acaulospora Gerd. & Trappe and Entrophospora R.N. Ames & R.W. Schneid., respectively (Gerdemann and Trappe 1974; Ames and Schneider 1979; Schüßler and Walker 2010).
As mentioned above, spores of Diversispora spp. arise terminally on a cylindrical or funnel-shaped subtending hypha, thus similarly to glomoid spores of Glomus macrocarpum Tul. & C. Tul., the type species of the genus Glomus Tul. & C. Tul. and the Glomeromycotina (Schüßler and Walker 2010). In Glomus spp., all layers of the subtending hyphal wall and the spore wall form concurrently and the subtending hyphal wall layer continuous with the structural laminate spore wall layer arises far below the spore base and, thereby, the subtending hypha is persistent and its wall is colored similarly to the laminate spore wall layer (Oehl et al. 2011b; Błaszkowski 2012). In contrast, the subtending hyphal wall of all Diversispora spp., also those with dark-colored spores, is hyaline to pale-colored and continuous only with a spore wall layer or layers overlying the main structural laminate spore wall layer, or the components of the subtending hyphal wall are continuous with all these spore wall layers (Oehl et al. 2011b). However, the laminate spore wall layer probably arises de novo sequentially after the full differentiation of the spore wall layer(s), which cover them, and starts developing either at the level of the lower surface of the spore wall layer directly adhering to its upper surface or slightly below the spore base (Oehl et al. 2011b). Thus, in the region of the subtending hyphal lumen, the laminate spore wall layer either is not present in the subtending hyphal wall at all or this layer is a small part of the subtending hyphal wall. The result is that the subtending hyphal wall layer continuous with the laminate spore wall layer does not create any or forms a weak physical support for the other subtending hyphal wall layer(s). Therefore, the spore subtending hypha of Diversispora spp. is usually fragile, and Oehl et al. (2011b) named spores of the genus diversisporoid.
Like in most species of the Glomeromycotina producing terminal, one-walled spores on a cylindrical or funnel-shaped sporogenous hypha, the spore wall structure of Diversispora spp. is simple and consists of two to four layers (Oehl et al. 2011b; Błaszkowski 2012). However, in contrast to many other genera of AMF forming spores in a similar way, spores of all Diversispora spp. do not react at all or rarely stain only faintly in Melzer’s reagent, except for Di. omaniana (Symanczik et al. 2014), which is the main focus of this study.
Although the spore morphological and histochemical features discussed above clearly allow to separate Diversispora spp. from its closest natural relatives, i.e., Corymbiglomus and Redeckera spp. (see Figs. 1, 2, and 3; Błaszkowski 2012), the differentiation between Diversispora spp. and some species of the genus Claroideoglomus C. Walker & A. Schüßler of the family Claroideoglomeraceae C. Walker & A. Schüßler may be difficult and uncertain (Oehl et al. 2011b; Błaszkowski, pers. observ.). This also applies to species of many other clades of the Glomeromycotina, mainly those producing spores similar in the mode of formation and construction to spores of Diversispora spp. Therefore, the identification and characterization of AMF have to be based on combined morphological data and those obtained from molecular phylogenetic analyses, as, for example, Gamper et al. (2009) suggested.
Based on our observations, of the molecular markers used in studies dealing with the identification and phylogeny of AMF, the small subunit gene (part; thereafter named SSU), the internal transcribed spacer 1 (ITS1), the 5.8S rRNA gene, ITS2 rDNA (full; hereafter named ITS), and the large subunit (part; thereafter named LSU) rRNA gene (thereafter together named SSU–ITS–LSU) is best suited for molecular and phylogenetic analyses of AMF because it is easily, successfully, and faultlessly (in 100% in our studies) amplified by the primers designed by Krüger et al. (2009), which, moreover, amplify the DNA fragment of all clades of the Glomeromycotina. The only drawback of sequences of the SSU–ITS–LSU segment is their weak species resolving power with respect to some Rhizoglomus spp., in which the variability of the ITS part may reach 20% (Stockinger et al. 2009). To overcome the problem, Stockinger et al. (2014) came back to earlier findings of Redecker and Raab (2006), which proved that the information contained in the largest subunit of RNA polymerase II (RPB1) gene is less variable and its species resolution is higher, and, consequently, constructed numerous primers to amplify the RPB1 gene of the Glomeromycotina. Our analyses of RPB1 sequences also showed that their variability did not exceed 0.5%. However, the use of the RPB1 gene in studies of AMF is much more difficult than that of the SSU–ITS–LSU nrDNA segment, which is further outlined in the Discussion section. Nonetheless, in phylogenetic studies, it is widely recognized that the study of several loci substantially improves the reliability and resolution power of phylogenies (James et al. 2006).
Currently, the genus Diversispora is represented by 15 species of confirmed molecular phylogeny and four species, which Oehl et al. (2011b) included into the genus based only on morphological characters. However, the literature data and molecular sequences deposited in public databases prove that the vast majority of AMF living in the world remain unnamed and many of them belong in the genus Diversispora (Gamper et al. 2009; Balázs et al. 2015; Błaszkowski et al. 2015b; Błaszkowski, pers. observ.).
Diversispora omaniana was described from spores formed singly at the top of a cylindrical to funnel-shaped sporogenous hypha (Symanczik et al. 2014). Phylogenetic analyses of molecular sequences of selected species of the Glomeromycotina placed the fungus in a sister clade to a clade grouping four Diversispora spp. Unfortunately, in the analyses, no sequence of Corymbiglomus and Redeckera spp. was considered because the morphology of members of both genera and the ecology of Redeckera spp. differed essentially from those of Di. omaniana (see the section Discussion regarding the taxa). However, subsequent pilot molecular phylogenetic analyses suggested that Di. omaniana does not belong in the genus Diversispora, but should represent a separate clade in the Diversisporales.
In addition, we grew, in single-species cultures, an AMF of morphological features, prompting that it is an undescribed member of the genus Diversispora, which also confirmed the pilot molecular analyses mentioned above.
Consequently, considering the information presented above, the aims of this study were to reconstruct the true phylogeny of the “omaniana” fungus and describe a new species in the genus Diversispora.
Materials and methods
Origin of study material, establishment and growth of trap and single-species cultures, extraction of spores, and staining of mycorrhizal structures
Trap and single-species cultures of the “omaniana” fungus were established and grown as described by Symanczik et al. (2014).
Spores of Di. sabulosa were first extracted from a pot trap culture inoculated with the rhizosphere soil and root fragments of Ammophila arenaria (L.) Link that had colonized maritime sand dunes of the Curonian Spit, Lithuania. The spores were subsequently used to establish single-species cultures. The trap and single-species cultures were established and grown as described by Błaszkowski et al. (2012), using Plantago lanceolata L. as the host plant. Spores for morphological and molecular analyses and roots for studies of mycorrhizal structures were collected as described by Błaszkowski et al. (2015a). Spores and roots came from five- to six-month-old cultures. Roots were stained following the protocol of Błaszkowski (2012).
Microscopy and nomenclature
At least 50–100 spores of each species mounted in water, lactic acid, polyvinyl alcohol/lactic acid/glycerol (PVLG; Omar et al. 1979), and a mixture of PVLG and Melzer’s reagent (1:1, v/v) were examined to determine their morphological features and the phenotypic and histochemical characters of spore wall layers. The preparation of spores and mycorrhizal structures for study and photography were as described previously (Błaszkowski 2012; Błaszkowski et al. 2012). Types of spore wall layers are those defined by Błaszkowski (2012), Stürmer and Morton (1997), and Walker (1983). Color names are from Kornerup and Wanscher (1983). Nomenclature of fungi and the authors of fungal names are from the Index Fungorum website (http://www.indexfungorum.org/AuthorsOfFungalNames.htm). Voucher specimens of the new species were mounted in PVLG and a mixture of PVLG and Melzer’s reagent (1:1, v/v) on slides and deposited at ETH in Zurich, Switzerland (Z + ZT; holotype), the Department of Ecology, Protection and Shaping of Environment (DEPSE), West Pomeranian University of Technology, Szczecin, and in the herbarium at Oregon State University (OSC) in Corvallis, Oregon, USA (isotypes).
Molecular phylogeny, DNA extraction, polymerase chain reaction, cloning, and DNA sequencing
Crude DNA of the “omaniana” fungus and the new Diversispora sp. was extracted from eight single spores of each fungus. Details of the treatment of the spores prior to polymerase chain reactions (PCRs), the conditions, and primers used in the PCRs to obtain SSU–ITS–LSU sequences were as those described in Błaszkowski et al. (2015a, b), Krüger et al. (2009), and Symanczik et al. (2014).
In order to obtain RPB1 sequences of the two fungi, nested PCRs were performed in conditions recommended and with primers designed by Stockinger et al. (2014). The first PCR was performed with the primers RPB1-DR160fmix (a,b,c,d) and RPB1-HS189GPf, and the second with RPB1-HS189GPf and RPB1-DR1210r. In the second, nested PCR, 0.5 μL of the first PCR product was used as the template. Cloning and sequencing of PCR products to obtain both types of sequences were performed as described by Błaszkowski et al. (2015a). The sequences were deposited in GenBank (MG459179–MG459217).
Sequence alignment and phylogenetic analyses
After pilot molecular phylogenetic analyses, which confirmed the supposition that the phylogenetic position of the species originally described as Di. omaniana is erroneous and the AMF found by us is an undescribed Diversispora sp., three sequence sets were assembled, two with sequences of the SSU–ITS–LSU nrDNA region and one with sequences of the RPB1 gene. Sequence identity matrices of the most closely related species to the “omaniana” fungus and the new Diversispora sp. were done using the BioEdit software (Hall 1999). All comparisons were performed on sequences of the same length.
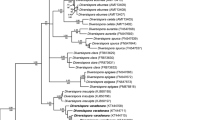
The first SSU–ITS–LSU sequence set was established to determine the taxonomic rank of the “omaniana” fungus among families of the Diversisporales sensu Walker and Schüßler (2004). Sixty-six sequences of the set characterized 26 species of seven families of the Diversisporales and G. macrocarpum of the family Glomeraceae in the order Glomerales J.B. Morton & Benny, which served as an outgroup (Fig. 1). Most sequences (28, 2–5 each) represented 11 species known to belong in the Diversisporaceae. The “omaniana” fungus was represented by seven sequences.
A 50% majority rule consensus phylogram inferred from a Bayesian analysis of SSU–ITS–LSU nrDNA sequences of Desertispora omaniana comb. nov., 25 other species of arbuscular mycorrhizal fungi (AMF) of the order Diversisporales, and Glomus macrocarpum as the outgroup. The new combination is in bold and followed by its GenBank accession numbers. The Bayesian posterior probabilities ≥ 0.50 and maximum likelihood (ML) bootstrap values ≥ 50% are shown near the branches, respectively. The bar indicates 0.05 expected change per site per branch
The second SSU–ITS–LSU sequence set mainly aimed at finding the phylogenetic position of the putative new Diversispora sp. among described species of the Diversispora clade. It was also supposed to support the conclusion of the existence of a new taxon represented by the “omaniana” fungus in the Diversisporales. The set consisted of 78 sequences of 13 certain Diversispora spp., the “omaniana” fungus, and the newly described Di. sabulosa, as well as C. corymbiforme Błaszk. & Chwat, three Redeckera spp., and Pacispora scintillans (S.L. Rose & Trappe) Sieverd. & Oehl ex C. Walker, Vestberg & A. Schüßler, of which the latter served as an outgroup taxon (Fig. 2). Each species was represented by 1–7 sequences. All sequences used in the alignment covered the whole SSU–ITS–LSU nrDNA segment amplified by the primers of Krüger et al. (2009), except the LSU sequences of Di. celata C. Walker, Gamper & A. Schüßler and the SSU–ITS sequences of R. fulvum (Berk. & Broome) C. Walker & A. Schüßler and R. pulvinatum (Henn.) C. Walker & A. Schüßler.
A 50% majority rule consensus phylogram inferred from a Bayesian analysis of SSU–ITS–LSU nrDNA sequences of Desertispora omaniana comb. nov., Diversispora sabulosa sp. nov., 18 other species of AMF of the order Diversisporales, and Pacispora scintillans as the outgroup. The new combination and new species are in bold and followed by their GenBank accession numbers. The Bayesian posterior probabilities ≥ 0.50 and ML bootstrap values ≥ 50% are shown near the branches, respectively. The bar indicates 0.1 expected change per site per branch
The RPB1 set contained 20 sequences of ten Diversispora spp., including the new Di. sabulosa, four sequences of the “omaniana” fungus, and 2–3 sequences each of representatives of the genera Corymbiglomus, Redeckera, and Acaulospora, with A. laevis being the outgroup taxon (Fig. 3).
A 50% majority rule consensus phylogram inferred from a Bayesian analysis of RPB1 sequences of Desertispora omaniana comb. nov., Diversispora sabulosa sp. nov., 22 other species of AMF of the order Diversisporales, and Glomus macrocarpum as the outgroup. The new combination and species are in bold and followed by their GenBank accession numbers. The Bayesian posterior probabilities ≥ 0.50 and ML bootstrap values ≥ 50% are shown near the branches, respectively. The bar indicates 0.1 expected change per site per branch
All sequence sets were aligned separately with MAFFT v.7 using the auto option (http://mafft.cbrc.jp/alignment/server). In the SSU–ITS–LSU sets, indels were coded by means of the simple indel coding algorithm (Simmons et al. 2001) as implemented in GapCoder (Young and Healy 2003), and these binary character sets were added to the nucleotide alignments, as described and justified by Błaszkowski et al. (2015c). The RPB1 set comprised only sequences of the RPB1 gene. Bayesian (BI) phylogenetic analyses of the SSU–ITS–LSU and RPB1 sequences were conducted with MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001). In all analyses, the GTR nucleotide substitution model was used, which was selected by jModelTest (Posada 2008), considering the selection of the Akaike information criterion. Four Markov chains were run for 5,000,000 generations, sampling every 100 steps, with a burn-in at 7500 sampled trees. Maximum likelihood (ML) phylogenetic analyses of sequences of the two loci were carried out with the raxmlGUI (Silvestro and Michalak 2012) implementation of RAxML (Stamatakis 2014), using the GTRGAMMA algorithm. Rapid bootstrap analysis with 1000 replicates was used to determine the support of the branches. In both BI and ML analyses of the SSU–ITS–LSU sequences, the sequence sets were divided into four partitions. The generated phylogenetic trees were visualized and edited in MEGA6 (Tamura et al. 2013).
Results
General data and phylogeny
To confirm the supposition of the earlier erroneously established phylogenetic position of the species originally described as Di. omaniana and, consequently, to give the fungus the appropriate status within the Glomeromycotina, as well as to determine the position of the putative undescribed Diversispora sp. among other Diversispora spp. of known molecular phylogeny, three sequence sets were assembled, two with sequences of the SSU–ITS–LSU nrDNA region and one with sequences of the RPB1 gene.
Bayesian and ML analyses of SSU–ITS–LSU sequences of the first sequence set with representatives of all families of the Diversisporales sensu Walker and Schüßler (2004) grouped the “omaniana” fungus in a separate clade at the rank of genus, whose sister clade contained sequences of C. corymbiforme and R. megalocarpum distributed separately in two subclades, each at the rank of genus (Fig. 1). The three generic clades connected a node with a large sister clade clustering sequences of Diversispora spp., O. bareae and T. nevadensis, which represented the Diversisporaceae clade, together with the “omaniana” fungus, C. corymbiforme, and R. megalocarpum. The Diversisporaceae clade, as well as the “omaniana” and C. corymbiforme clades, were fully supported in both analyses (BI = 1.0, ML = 100%). The supports of the R. megalocarpum clade and the node connecting the C. corymbiforme and R. megalocarpum clades with the “omaniana” clade were also full and strong. The alignment of the sequence set had a length of 2057 characters, of which 72.2% were phylogenetic informative. The identity values of the seven SSU–ITS–LSU sequences of the “omaniana” fungus calculated with BioEdit (Hall 1999) were 96.7–99.4%.
Bayesian and ML analyses of the second set with SSU–ITS–LSU sequences confirmed the novelty of the Diversispora sp. tested in this study and placed the species in an own clade positioned below a clade with Di. aurantia (Błaszk. et al.) C. Walker & A. Schüßler and D. spurca distributed in two subclades and above a clade with three monophyletic groups represented by Di. gibbosa, Di. peridiata Błaszk. et al., and Di. trimurales (Koske & Halvorson) C. Walker & A. Schüßler (Fig. 2). The Di. sabulosa clade was fully (BI = 1.0) and highly (ML = 99%) supported in the analyses. The identity values of the five SSU–ITS–LSU sequences of Di. sabulosa ranged from 98.0% to 99.8%.
Also, the analyses of the second sequence set proved that the basal AMF group of the Diversisporaceae consists of two sister genera Corymbiglomus and Redeckera, and a new genus represented by the “omaniana” fungus (Fig. 2). The “omaniana” clade was fully supported in both BI and ML analyses. The large clade C. corymbiforme/Redeckera spp./“omaniana”, as well as the clades C. corymbiforme only and Redeckera spp. only, also obtained full or high support values. Importantly, the node connecting the C. corymbiforme/Redeckera spp./“omaniana” clade with the clade comprising all Diversispora spp. of known molecular phylogeny, and, thereby, representing the family Diversisporaceae, was also fully supported in both analyses. The alignment had a length of 1927 characters, of which 80.38% were phylogenetic informative.
In the tree with RPB1 sequences, the new Di. sabulosa was also located in a separate clade, fully supported in BI and ML analyses, but the clade was embedded between a clade with Di. arenaria and Di. jakucsiae distributed in two well-supported subclades and a fully supported clade with Di. slowinskiensis (Fig. 3). The positions of the “omaniana” clade and the clades with C. corymbiforme and R. fulvum relative to each other and the other analyzed members of the Diversisporaceae were identical to the positions of the taxa visualized in the SSU–ITS–LSU trees (Figs. 1 and 2). In both BI and ML analyses, the “omaniana” clade received full support values. The clades with C. corymbiforme and R. fulvum were fully or very strongly supported, and the support values of the Diversisporaceae clade were 1.0 and 71% in BI and ML analyses, respectively. The length of the alignment was 829 characters, of which 71.41% were phylogenetic informative. The identity values of the two RPB1 sequences of Di. sabulosa and the four RPB1 sequences of the “omaniana” fungus were 99.8% and 99.3–99.8%, respectively.
The identities of the SSU–ITS–LSU sequences of “omaniana” vs. C. corymbiforme, “omaniana” vs. R. megalocarpum, and C. corymbiforme vs. R. megalocarpum calculated with BioEdit (Hall 1999) were 79.7–81.8%, 83.0–83.8%, and 86.4–87.6%, respectively. Of the Diversispora spp. most closely related to Di. sabulosa sp. nov. (see above and Fig. 2), Di. aurantia had the highest identity of the SSU–ITS–LSU sequences: 92.2–92.7%.
The identities of the RPB1 sequences of “omaniana” vs. C. corymbiforme, “omaniana” vs. R. fulvum, and C. corymbiforme vs. R. fulvum were 84.6–84.9%, 81.4–82.1%, and 92.0–92.5%, respectively. The identities of the RPB1 sequences of the new Diversispora sp. and its closest relatives Di. arenaria, Di. jakucsiae, and Di. slowinskiensis (Fig. 3) ranged from 96.1% (Di. arenaria) to 96.7% (Di. jakucsiae).
Thus, the phylogenetic analyses discussed above unambiguously proved that the “omaniana” fungus should represent a new taxon at the rank of genus belonging to the family Diversisporaceae and the AMF found by us is a new species in the genus Diversispora. Therefore, the taxa are described below as Desertispora gen. nov., with De. omaniana comb. nov., and Di. sabulosa sp. nov.
Taxonomy
Erection of a new genus
Desertispora Błaszk., Kozłowska, Ryszka, Al-Yahya’ei & Symanczik, gen. nov., MB 823563. Figs. 4, 5
Spores of Desertispora omaniana. 4. Spore wall layers (swl) 1–3. 5. Subtending hyphal wall layers (shwl) 1–3 continuous with spore wall layers (swl) 1–3 and septum (s) in the lumen of the subtending hypha. 4. Spore in PVLG + Melzer’s reagent. 5. Spore in PVLG. 4, 5. Differential interference microscopy. Bars: 4, 5 = 10 μm
Type species: Desertispora omaniana (Symanczik, Błaszk. & Al-Yahya’ei) Symanczik, Błaszk., Kozłowska & Al-Yahya’ei.
Etymology: Desertispora, referring to the desert habitat in which the fungus was originally found.
Diagnosis: Differs from other genera in the Diversisporaceae and Diversisporales, whose species form spores at the tip of a cylindrical or funnel-shaped sporogenous hypha, in the possession of a spore wall with a structural laminate layer reacting in Melzer’s reagent, and in having the specific sequences of the nrDNA regions of ITS1: GAAATTTAATTCCCGGGAATTCTTGGTTCCCG, ITS2: AAAATGAATTCTGGTTCAAGTCAAAAACGTTCTATGC and TCATTATGTTCCCCACCT, and the LSU gene: CGAAGTTCCAATCAGATACGCTCTCG and ACATTCGTGGGTCTTGACGACCACGGGTTAGAGCGTTC, as well as the RPB1 gene: for example, TCTCTTCTCTTCCCTCTCTCTTCTCTTCCTCTTCC, CAAATTCACTCATGTTTTCATTATTACTTATTCATATCACGCGGGAAAAAT, and CTCCACCCACCCCGGAGCCAAGTACGTGATTAGGGATACCGGTGAAAGGATCGAC.
Description: Spores formed singly in soil, develop blastically at the tip of sporogenous hyphae. Spores with one subtending hypha. Spore wall composed of three layers (layers 1–3). Layer 1 evanescent, roughened, hyaline. Layer 2 permanent, pliable, uniform (not composed of sublayers), smooth, hyaline to pastel yellow (2A4). Layer 3 laminate, smooth, hyaline. In Melzer’s reagent, layer 1 always remains nonreactive, layer 2 usually darkens to pale yellow (3A3), and layer 3 usually stains pinkish (9A2) to dull red (11C3). Subtending hypha cylindrical to funnel-shaped, rarely constricted at the spore base, not breaking up in crushed spores. Wall of subtending hypha composed of three layers continuous with spore wall layers 1–3. Pore open or occluded by a curved septum continuous with some innermost laminae of spore wall layer 3; septum positioned up to 15.7 μm below the spore base. Spore contents of a hyaline to brownish yellow (5C8), sticky, opaque substance.
Desertispora omaniana (Symanczik, Błaszk. & Al-Yahya’ei) Symanczik, Błaszk., Kozłowska & Al-Yahya’ei, comb. nov., MB 823564. Figs. 4, 5
Basionym: Diversispora omaniana Symanczik, Błaszk. & Al-Yahya’ei. Mycologia 106: 247, 2014.
Spores formed singly in soil, develop blastically at the tip of sporogenous hyphae. Spores hyaline to brownish yellow (5C8); globose to subglobose; (85–)136(−170) μm diam.; rarely egg-shaped; 110–160 × 130–180 μm; with one subtending hypha. Spore wall composed of three layers (layers 1–3; Figs. 4, 5). Layer 1 evanescent, roughened, hyaline, (1.0–)1.9(−2.8) μm thick when intact, usually more or less deteriorated in mature spores, frequently partly or completely sloughed in older specimens. Layer 2 permanent, pliable, uniform (not composed of sublayers), smooth, hyaline to pastel yellow (2A4), (1.5–)2.4(−3.8) μm thick. Layer 3 laminate, smooth, hyaline, (4.8–)8.9(−22.0) μm thick, frequently stratifying into groups of or single laminae in crushed spores. In Melzer’s reagent, layer 1 always remains nonreactive, layer 2 usually darkens to pale yellow (3A3), and layer 3 usually stains pinkish (9A2) to dull red (11C3; Fig. 4). Subtending hypha hyaline to pastel yellow (2A4); cylindrical to funnel-shaped, rarely constricted at the spore base; (10.8–)14.3(−23.5) μm wide at the spore base, not breaking up in crushed spores (Fig. 5). Wall of subtending hypha hyaline to pastel yellow (2A4); (2.3–)4.2(−5.5) μm thick at the spore base; composed of three layers continuous with spore wall layers 1–3 (Fig. 5). Pore (3.0–)7.1(−14.3) μm diam., open or occluded by a curved septum, 1.0–1.3 mm thick, continuous with some innermost laminae of spore wall layer 3; septum positioned up to 15.7 μm below the spore base (Fig. 5). Spore content of a hyaline to brownish yellow (5C8), sticky, opaque substance (Figs. 4, 5).
Specimens examined: Holotype: 3222 (DEPSE), isotypes: 3221, 3223–3243, and two slides at OSC (Symanczik et al. 2014).
Distribution and habitat: To date, found only based on spores extracted from trap cultures inoculated with the rhizosphere soils of Salvadora persica Wall., Tetraena qatarense Beier & Thulin. and unrecognized grasses, in undisturbed natural field located at Al-Kamel in the Al-Sharqyia region of Oman (22°14′911″N, 59°10′53″E; Symanczik et al. 2014). BLAST queries did not show any SSU–ITS–LSU sequence of identity at least 97% to the SSU–ITS–LSU sequences of De. omaniana. The highest identity of the RPB1 sequences of De. omaniana was found in comparison with the RPB1 HG315981 sequence of Di. epigaea, which was only 86%.
Description of a new species
Diversispora sabulosa Błaszk. & Kozłowska, sp. nov., MB 823568. Figs. 6–13
Spores and mycorrhizal structures of Diversispora sabulosa. 6. Intact spores. 7–9. Spore wall layers (swl) 1 and 2. 10, 11. Spore wall layers (swl) 1 and 2 continuous with wall layers (shwl) 1 and 2 of the cylindrical subtending hypha and a septum (s) enclosing its lumen. 12. Spore wall layers (swl) 1 and 2 continuous with wall layers (shwl) 1 and 2 of the subtending hypha constricted at the spore base. 13. Arbuscules (a) with a trunk (t) and intraradical hypha (ih). 6–8, 10, 13. Spores in PVLG. 9, 11, 12. Spores in PVLG + Melzer’s reagent. 6–13. Differential interference microscopy. Bars: 6 = 50 μm, 7–13 = 10 μm
Holotype: ZT Myc 58,675 (Z + ZT), isotypes: 3593–3601 (DEPSE), and OSC 161509, OSC 161510 (OSC).
Etymology: Latin, sabulosa, referring to the sandy habitat in which the fungus exists.
Sporocarps unknown. Spores formed singly in soil (Figs. 6–12); arise blastically at the tip of sporogenous hyphae developed from mycorrhizal extraradical hyphae. Spores deep yellow (4A8) to orange yellow (4B8), rarely brownish yellow (5C8); globose to subglobose; (17–)51(−67) μm diam.; frequently ovoid; 80–110 × 95–130 μm; with one subtending hypha (Figs. 6–12). Spore wall consists of two layers (Figs. 7–12). Layer 1, forming the spore surface, uniform (not divided into visible sublayers), permanent, semi-flexible, hyaline to light yellow (4A5), (1.2–)2.5(−3.6) μm thick, usually tightly adherent to the upper surface of layer 2, sometimes separating from it, especially in vigorously crushed spores (Figs. 7–12). Layer 2 laminate, permanent, smooth, deep yellow (4A8) to orange yellow (4B8), rarely brownish yellow (5C8), (3.8–)5.2(−7.5) μm thick, consisting of very thin, < 0.5 μm thick, laminae, tightly adherent to each other (Figs. 7–12). Layers 1 and 2 do not show amyloid or dextrinoid reaction in Melzer’s reagent (Figs. 9, 11, 12). Subtending hypha hyaline to pale yellow (3A3), rarely light yellow (4A4); straight or recurved, cylindrical to slightly funnel-shaped, sometimes constricted at the spore base; (5.6–)6.9(−8.5) μm wide at the spore base (Figs. 10–12). Wall of subtending hypha hyaline to pale yellow (3A3), rarely light yellow (4A4); (1.6–)2.5(−4.3) μm thick at the spore base; continuous with spore wall layers 1 and 2 (Figs. 10–12). Pore (1.4–)2.4(−3.0) μm diam., open or occluded by a straight or curved septum, formed by subtending hyphal wall layer 2; septum positioned up to 5.4 μm below the spore base (Figs. 10–12). Germination unknown.
Mycorrhizal associations: The presence of spores of Di. sabulosa in a trap culture inoculated with the rhizosphere soil of Ammophila arenaria (L.) Link suggested that the AMF forms a symbiotic association with the plant. However, no molecular analysis was performed on its roots to confirm the supposition. The field soil × root sample was collected on 31 September 2013.
In single-species cultures with P. lanceolata as the host plant, Di. sabulosa formed mycorrhiza with arbuscules and intraradical and extraradical hyphae (Fig. 13). No vesicles were found. Arbuscules were uniformly distributed along the root fragments examined (Fig. 13). Intraradical hyphae were straight, slightly recurved, or formed Y-shaped branches and coils (Fig. 13). Extraradical hyphae occurred infrequently or abundantly, depending on the root fragment examined. In 0.1% Trypan blue, all of the structures stained faintly [violet white (15A2) to pale violet (15A3); Fig. 13].
Distribution and habitat: Despite that J. Błaszkowski personally examined the occurrence of AMF in ca. 2500 field-collected rhizosphere soil samples and ca. 3000 trap cultures inoculated with cultivated and natural soils collected in different regions of Africa, Asia, Brazil, Europe, and the USA, Di. sabulosa was found only in one trap culture representing mobile maritime dunes of the Curonian Spit located in the north of Lithuania (55°26′N, 21°04′E). The dunes are the highest sand dunes in Europe: their average height is 35 m, but some attain a height of 60 m (https://en.wikipedia.org/wiki/Curonian_Spit). Since 2000, the Curonian Spit has been on UNESCO’s World Heritage List under cultural criteria “V”. The vegetation on the Curonian Spit is varied and unique due to the specific climatic conditions of the sea coast (http://daugenis.mch.mii.lt/apsamogitia/LANKYTINOS_VIETOS/nerija.en.htm). About 700 plant species were found on the spit, among which the prevailing plants are those well adapted to infertile and unstable sands, such as, for example, A. arenaria.
BLAST queries confirmed the rare occurrence of Di. sabulosa in the world determined on the basis of studies of the presence of spores of AMF in field soils and trap cultures: in public databases, there is no sequence of the SSU–ITS–LSU nrDNA segment and the RPB1 gene of identity ≥ 97%.
Discussion
The investigations discussed in this paper confirmed our suppositions that the AMF originally described as Di. omaniana does not belong in the genus Diversispora, but represents a new taxon in the family Diversisporaceae (Figs. 1, 2, and 3), which was described here as Desertispora gen. nov. These studies also proved that the putative undescribed Diversispora sp. that we found in maritime sand dunes of the Curonian Spit, Lithuania, is a new species of the genus (Figs. 2 and 3), here named Di. sabulosa.
Despite that the features of De. omaniana, especially the histochemical traits of its spores, do not fit the characters defining the genus Diversispora and other clades of the order Diversisporales comprising species producing spores terminally on a cylindrical or funnel-shaped sporogenous hypha (Oehl et al. 2011b; Schüßler and Walker 2010; Symanczik et al. 2014) and Di. sabulosa in morphology is unique compared to the other Diversispora spp., the certain determination of the phylogenetic position of the two species within the Diversisporales was possible only after molecular phylogenetic analyses of their sequences. The analyses proved that the new genus Desertispora is most closely related to the genera Corymbiglomus and Redeckera and showed the closest relatives of Di. sabulosa (Figs. 1, 2, and 3).
Morphologically, De. omaniana differs clearly from Corymbiglomus spp. and Redeckera spp. Corymbiglomus corymbiforme, whose sequences were used in the molecular phylogenetic analyses discussed here (Figs. 1, 2, and 3), forms spores mainly in corymbiform clusters, rarely singly (Błaszkowski 1995, 2012), whereas spores of De. omaniana arise only singly (Figs. 4, 5; Symanczik et al. 2014). Moreover, C. corymbiforme spores are surrounded by a hyphal mantle, their spore wall layer 3 is flexible to semi-flexible, and none of the three spore wall layers stains in Melzer’s reagent. In De. omaniana, spores are naked (not covered by hyphae; Figs. 4, 5), and their laminate spore wall 3 usually stains pinkish (9A2) to dull red (11C3) in Melzer’s reagent (Fig. 4). Spores of C. globiferum (Koske & C. Walker) Błaszk. & Chwat and C. tortuosum (N.C. Schenck & G.S. Sm.) Błaszk. & Chwat are also covered with a peridium, do not react in Melzer’s reagent, and their spore wall is one-layered (C. tortuosum) or three-layered with a semi-flexible layer 3 (C. globiferum; Koske and Walker 1986; Błaszkowski 2012). The last of the described Corymbiglomus spp., C. pacificum Oehl et al., produces single naked spores, thus similarly as De. omaniana does (Figs. 4, 5), but spores of the former species have a more complex subcellular structure, which, according to Medina et al. (2014), consists of two walls, each with three layers.
Similarly as in comparison to C. corymbiforme, De. omaniana differs substantially from Redeckera spp. All of the known Redeckera spp. have been found to form spores mainly in epigeous compact sporocarps with a peridium (Gerdemann and Trappe 1974; Redecker et al. 2007), rarely singly (Oehl et al. 2011b). The ability to form mycorrhiza by Redeckera spp. remains unknown because many attempts to grow them in culture have failed (Redecker et al. 2007). The spore wall of Redeckera spp. consists of only one laminate layer not reacting in Melzer’s reagent. Their spore subtending hypha, whose wall is one-layered, is usually fragile, inflated slightly below the spore base, and its lumen is closed by a straight, broad septum positioned at the level of the spore base; it is rarely open (Gerdemann and Trappe 1974; Redecker et al. 2007; Oehl et al. 2011b; Błaszkowski 2012). In contrast, De. omaniana produced only hypogeous ectocarpic spores (Figs. 4, 5) and the species was grown in single-species cultures, which were established from only one spore (Symanczik et al. 2014). The spore wall of De. omaniana comprises three layers (Figs. 4, 5), of which the laminate layer 3 usually stains in Melzer’s reagent (Fig. 4). The subtending hypha of De. omaniana is persistent, consists of three layers continuous with spore wall layers 1–3, is cylindrical to funnel-shaped without an inflation, and its lumen is closed by a curved septum positioned up to 15.7 μm below the spore base (Fig. 5); in young specimens, it is frequently open. Probably, the only characters uniting De. omaniana and Redeckera spp. are those of the spore content. In De. omaniana (Figs. 4, 5) and, for example, in R. fulvum and R. megalocarpum, the spore content is colorless to yellow-colored, sticky, and opaque (Błaszkowski 2012).
Apart from the clear morphological differences discussed above, De. omaniana, Corymbiglomus spp., and Redeckera spp. are separated by a large molecular distance (see the above section entitled General data and phylogeny; Błaszkowski 2012; Błaszkowski and Chwat 2013).
The most distinctive morphological character of Di. sabulosa is its spore wall layer 1, which is permanent, uniform, usually colorless, and relatively thick when compared with the laminate spore wall layer 2 (Figs. 7–12).
The separateness of Di. sabulosa unambiguously proved the performed molecular phylogenetic analyses, particularly those of sequences of the SSU–ITS–LSU nrDNA segment, which considered all Diversispora spp. of known natural phylogeny. The analyses clustered Di. sabulosa sequences in a separate clade, nested between a clade with Di. aurantia and Di. spurca, and a clade with sequences of Di. gibbosa, Di. peridiata, and Di. trimurales (Fig. 2). However, of the five Diversispora spp., only Di. peridiata produces spores with a two-layered spore wall and the two layers are permanent (Błaszkowski et al. 2015b). Several morphological characters clearly distinguish Di. sabulosa from Di. peridiata. Apart from single spores, Di. peridiata produces spores in clusters and the spores are occasionally covered with peridium-like hyphae. Diversispora sabulosa forms only single spores without a peridium (Figs. 6–12). Spores of Di. peridiata are 1.5–2.1-fold larger when globose, and their spore wall layer 1 and the subtending hyphal wall at the spore base are 1.5–1.7-fold and 1.4–2.8-fold thicker, respectively. In addition, spores of the species are lighter [yellowish white (4A2) to maize yellow (4A6)], and their spore wall layer 1 may be much darker [brownish yellow (5C8)] and is never as hyaline as in D. sabulosa (Figs. 7–12).
The BI and ML phylogenetic analyses with sequences of the RPB1 gene also located Di. sabulosa in a separate, fully supported clade, but the clade was embedded between a clade with Di. arenaria and Di. jakucsiae distributed in two subclades and a clade with Di. slowinskiensis (Fig. 3), which, in the SSU–ITS–LSU tree, were placed far from the new species (Fig. 2). However, these relationships need to be treated with caution because the RPB1 analyses did not consider sequences of Di. aurantia and Di. spurca included in the SSU–ITS–LSU analyses (Figs. 1 and 2); many attempts to obtain them failed. Nevertheless, morphologically, Di. sabulosa differs substantially from Di. arenaria, Di. jakucsiae, and Di. slowinskiensis, for example, in the spore wall structure, which is two-layered in the former new AMF (Figs. 7–12), and three-layered in the three latter species (Błaszkowski et al. 2001, 2015b; Balázs et al. 2015). The molecular distance between the species is also large (see the above section entitled General data and phylogeny).
As mentioned in the Introduction section, the use of the RPB1 gene in studies of AMF still encounters many limitations. Unfortunately, the primers designed by Stockinger et al. (2014) do not amplify all clades of the subphylum, and many of them either amplify only short or much shorter than expected fragments of the RPB1 gene, such as, for example, those recommended for Diversispora spp., or do not work at all (pers. observ.). However, the limited usability of the primers mainly results from that they amplify only particular and not all clades of the Glomeromycotina. Thus, the recognition of molecular properties of targeted AMF based on the RPB1 gene has to be preceded by the identification of their position within the subphylum, using, for example, the primers by Krüger et al. (2009).
References
Ames RN, Schneider RW (1979) Entrophospora, a new genus in the Endogonaceae. Mycotaxon 8:347–352
Balázs TK, Błaszkowski J, Chwat G, Góralska A, Gáspár BK, Lukács AF, Kovács GM (2015) Spore-based study of arbuscular mycorrhizal fungi of semiarid sandy areas in Hungary, with Diversispora jakucsiae sp. nov. Mycol Prog 14:1021. https://doi.org/10.1007/s11557-014-1021-z
Błaszkowski J (1995) Glomus corymbiforme, a new species in Glomales from Poland. Mycologia 87:732–737. https://doi.org/10.2307/3760819
Błaszkowski J (2012) Glomeromycota. W. Szafer Institute of Botany, Polish Academy of Sciences, Kraków
Błaszkowski J, Chwat G (2013) Septoglomus deserticola emended and new combinations in the emended definition of the family Diversisporaceae. Acta Mycol 48(1):89–103. https://doi.org/10.5586/am.2013.011
Błaszkowski J, Tadych M, Madej T (2001) Glomus arenarium, a new species in Glomales (Zygomycetes). Acta Soc Bot Pol 70:97–101. https://doi.org/10.5586/asbp.2001.013
Błaszkowski J, Kovács GM, Gáspár BK, Balázs TK, Buscot F, Ryszka P (2012) The arbuscular mycorrhizal Paraglomus majewskii sp. nov. represents a distinct basal lineage in Glomeromycota. Mycologia 104:148–156. https://doi.org/10.3852/10-430
Błaszkowski J, Chwat G, Symanczik S, Góralska A (2015a) Dominikia duoreactiva sp. nov. and Dominikia difficilevidera sp. nov., two new species in the Glomeromycota. Botany 93:389–396. https://doi.org/10.1139/cjb-2015-0016
Błaszkowski J, Furrazola E, Chwat G, Góralska A, Lukács AF, Kovács GM (2015b) Three new arbuscular mycorrhizal Diversispora species in Glomeromycota. Mycol Prog 14:105. https://doi.org/10.1007/s11557-015-1122-3
Błaszkowski J, Chwat G, Góralska A, Ryszka P, Kovács GM (2015c) Two new genera, Dominikia and Kamienskia, and D. disticha sp. nov. in Glomeromycota. Nova Hedwigia 100:225–238. https://doi.org/10.1127/nova_hedwigia/2014/0216
Castillo CG, Borie F, Oehl F, Sieverding E (2016) Arbuscular mycorrhizal fungi biodiversity: prospecting in Southern-Central zone of Chile. A review. J Soil Sci Plant Nutr 16:400–422. https://doi.org/10.4067/S0718-95162016005000036
Gamper HA, Walker C, Schüßler A (2009) Diversispora celata sp. nov: molecular ecology and phylotaxonomy of an inconspicuous arbuscular mycorrhizal fungus. New Phytol 182:495–506. https://doi.org/10.1111/j.1469-8137.2008.02750.x
Gerdemann JW, Trappe JM (1974) The Endogonaceae in the Pacific Northwest. Mycol Mem 5:1–76
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. https://doi.org/10.1093/bioinformatics/17.8.754
James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ et al (2006) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443:818–822. https://doi.org/10.1038/nature05110
Kornerup A, Wanscher JH (1983) Methuen handbook of colour, 3rd edn. E. Methuen, London
Koske RE, Walker C (1986) Glomus globiferum: a new species of Endogonaceae with a hyphal peridium. Mycotaxon 26:133–142
Krüger M, Stockinger H, Krüger C, Schüßler A (2009) DNA-based species level detection of Glomeromycota: one PCR primer set for all arbuscular mycorrhizal fungi. New Phytol 183:212–223. https://doi.org/10.1111/j.1469-8137.2009.02835.x
Medina J, Cornejo P, Borie F, Meier S, Palenzuela J, Vieira HEE, Ferreira ACA, da Silva GA, Sánchez-Castro I, Oehl F (2014) Corymbiglomus pacificum, a new glomeromycete from a saline lakeshore in Chile. Mycotaxon 127:173–183. https://doi.org/10.5248/127.173
Oehl F, da Silva GA, Goto BT, Maia LC, Sieverding E (2011a) Glomeromycota: two new classes and a new order. Mycotaxon 116:365–379. https://doi.org/10.5248/116.365
Oehl F, da Silva GA, Goto BT, Sieverding E (2011b) Glomeromycota: three new genera and glomoid species reorganized. Mycotaxon 116:75–120. https://doi.org/10.5248/116.75
Oehl F, da Silva GA, Sánchez-Castro I, Goto BT, Maia LC, Vieira HEE, Barea J-M, Sieverding E, Palenzuela J (2011c) Revision of Glomeromycetes with entrophosporoid and glomoid spore formation with three new genera. Mycotaxon 117:297–316. https://doi.org/10.5248/117.297
Omar MB, Bolland L, Heather WA (1979) A permanent mounting medium for fungi. Bull Br Mycol Soc 13:31–32. https://doi.org/10.1016/S0007-1528(79)80038-3
Pagano MC, Oehl F, Silva GA, Maia LC, Silva DK, Cabello MN (2016) Advances in arbuscular mycorrhizal taxonomy. In: Pagano M (ed) Recent advances on mycorrhizal fungi. Springer International Publishing, Switzerland, pp 15–21. https://doi.org/10.1007/978-3-319-24355-9_2
Palenzuela J, Ferrol N, Boller T, Azcón-Aguilar C, Oehl F (2008) Otospora bareai, a new fungal species in the Glomeromycetes from a dolomitic shrub land in Sierra de Baza National Park (Granada, Spain). Mycologia 100:296–305. https://doi.org/10.1080/15572536.2008.11832484
Palenzuela J, Barea JM, Ferrol N, Azcón-Aguilar C, Oehl F (2010) Entrophospora nevadensis, a new arbuscular mycorrhizal fungus from Sierra Nevada National Park (southeastern Spain). Mycologia 102(3):624–632. https://doi.org/10.3852/09-145
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. https://doi.org/10.1093/molbev/msn083
Redecker D, Raab PA (2006) Phylogeny of the Glomeromycota (arbuscular mycorrhizal fungi): recent developments and new gene markers. Mycologia 98:885–895. https://doi.org/10.1080/15572536.2006.11832618
Redecker D, Raab PA, Oehl F, Camacho FJ, Courtecuisse R (2007) A novel clade of sporocarp-forming species of glomeromycotan fungi in the Diversisporales lineage. Mycol Prog 6:35–44. https://doi.org/10.1007/s11557-007-0524-2
Redecker D, Schüßler A, Stockinger H, Stürmer SL, Morton JB, Walker C (2013) An evidence-based consensus for the classification of arbuscular mycorrhizal fungi (Glomeromycota). Mycorrhiza 23:515–531. https://doi.org/10.1007/s00572-013-0486-y
Schüßler A, Walker C (2010) The Glomeromycota. A species list with new families and new genera. Royal Botanic Garden Edinburgh, Gloucester
Silvestro D, Michalak I (2012) raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 12:335–337. https://doi.org/10.1007/s13127-011-0056-0
Simmons MP, Ochoterena H, Carr TG (2001) Incorporation, relative homoplasy, and effect of gap characters in sequence-based phylogenetic analyses. Syst Biol 50:454–462. https://doi.org/10.1080/106351501300318049
Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, Bonito G, Corradi N, Grigoriev I, Gryganskyi A, James TY, O’Donnell K, Roberson RW, Taylor TN, Uehling J, Vilgalys R, White MM, Stajich JE (2016) A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108:1028–1046. https://doi.org/10.3852/16-042
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Stockinger H, Walker C, Schüßler A (2009) ‘Glomus intraradices DAOM197198’, a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol 183:1176–1187. https://doi.org/10.1111/j.1469-8137.2009.02874.x
Stockinger H, Peyret-Guzzon M, Koegel S, Bouffaud M-L, Redecker D (2014) The largest subunit of RNA polymerase II as a new marker gene to study assemblages of arbuscular mycorrhizal fungi in the field. PLoS One 9(10):e107783. https://doi.org/10.1371/journal.pone.0107783
Stürmer SL, Morton JB (1997) Developmental patterns defining morphological characters in spores of four species in Glomus. Mycologia 89:72–81. https://doi.org/10.2307/3761174
Symanczik S, Błaszkowski J, Chwat G, Boller T, Wiemken A, Al-Yahya’ei MN (2014) Three new species of arbuscular mycorrhizal fungi discovered at one location in a desert of Oman: Diversispora omaniana, Septoglomus nakheelum and Rhizophagus arabicus. Mycologia 106(2):243–259. https://doi.org/10.3852/106.2.243
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Walker C (1983) Taxonomic concepts in the Endogonaceae; spore wall characteristics in species descriptions. Mycotaxon 18:443–455
Walker C, Schüßler A (2004) Nomenclatural clarifications and new taxa in the Glomeromycota Pacispora. Mycol Res 108:981–982. https://doi.org/10.1017/S0953756204231173
Young ND, Healy J (2003) GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinformatics 4:6. https://doi.org/10.1186/1471-2105-4-6
Acknowledgements
This study was supported in part by the Polish National Centre of Science, grant nos. 2012/05/B/NZ8/00498 and 2012/07/N/NZ8/02363, which are greatly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Marco Thines
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Symanczik, S., Al-Yahya’ei, M.N., Kozłowska, A. et al. A new genus, Desertispora, and a new species, Diversispora sabulosa, in the family Diversisporaceae (order Diversisporales, subphylum Glomeromycotina). Mycol Progress 17, 437–449 (2018). https://doi.org/10.1007/s11557-017-1369-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-017-1369-y