Abstract
Purpose
Monitoring and predicting the cognitive state of subjects with neurodegenerative disorders is crucial to provide appropriate treatment as soon as possible. In this work, we present a machine learning approach using multimodal data (brain MRI and clinical) from two early medical visits, to predict the longer-term cognitive decline of patients. Using transfer learning, our model can be successfully transferred from one neurodegenerative disease (Alzheimer’s) to another (Parkinson’s).
Methods
Our model is a Deep Neural Network with siamese sub-modules dedicated to extracting features from each modality. We pre-train it with data from ADNI (Alzheimer’s disease), then transfer it on the smaller PPMI dataset (Parkinson’s disease). We show that, even when we do not fine-tune the filters learnt from the ADNI MRIs, the transferred model’s results are satisfying on PPMI.
Results
The first main result is that our model provides satisfying long-term predictions of cognitive decline from any pair of early visits, with no fixed time delay between these visits (provided the potential decline has started at the second visit). The second main result is that the prediction performance on Parkinson’s dataset (PPMI) reaches an AUC of 0.81 on PPMI after transfer learning from Alzheimer’s dataset (ADNI), without even having to re-train the image filters, versus an AUC of 0.72 for the model trained from scratch on PPMI.
Conclusions
First, our model is effective for predicting long-term cognitive decline from only two visits, even with irregular intervals of time. When dealing with neurodegenerative diseases, where patients often miss some control visits, this is an important finding. Second, our model is able to transfer the knowledge learnt from one neurodegenerative disease (Alzheimer’s) to another (Parkinson’s), when using the same imaging modalities (brain MRI) and different clinical variables. This makes it usable even for diseases that are rare or under-studied.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
There are some longitudinal studies where people at risk of developing neurodegenerative diseases are monitored during several years. These studies allow medical doctors and data scientists to gather medical records information and medical imaging from different sources. However, the relatively small size of bio-medical cohorts can induce over-fitting when using deep learning models, therefore hindering their effectiveness and generalization capacity. Moreover, due to technical hazards and patients’ life uncertainties, data collection can be irregular an incomplete, especially in the case of neurodegenerative diseases.
In this work, we first present a deep learning model able to use multimodal medical data from two arbitrarily chosen, early medical visits to predict longer-term cognitive decline in patients with suspected Alzheimer’s disease. Then, once this model is validated, we show that our model is adaptable, in the sense that, once pre-trained for a given disease, its knowledge can easily be transferred to another disease, even in the presence of a smaller dataset.
In short, our model has the following characteristics:
-
1.
It uses data from multiple modalities: medical images (brain MRI), clinical data, and risk factors associated with the pathology.
-
2.
Instead of being based on projections of the structural 3D MRI onto some 2D plane (leading to a loss of information), or on the (possibly noisy) extractions of some Regions of Interest (ROI), it is able to process directly the 3D MRI images by learning 3D filters.
-
3.
It takes as input two arbitrarily chosen medical visits, \(T_i\) and \(T_i+\delta \) (without strong constraints on the initial time \(T_i\) nor the time interval \(\delta \) between the two, contrarily to our previous model), and returns the predicted longer-term class (“stable” or “declining”). We observe very satisfying results with a potential decline observed between the two visits.
-
4.
It is adaptable, in the sense that it can be pre-trained for a given neurodegenerative disease and then its knowledge can be easily transferred to another neurodegenerative disease (using a target dataset with similar modality types, but fewer patients and/or visits). As a proof of concept, we use the case study of transfer learning from Alzheimer’s disease to Parkinson’s disease, with the same imaging modality (3D MRI) and similar risk factors, but different clinical variables. We will show that, even if we do not fine-tune the filters learnt on the images, it still gives very good results on the transferred dataset. This shows that our model could be easily re-used for a variety of neurodegenerative diseases which are too rare or under-studied to have dedicated labelled bulky datasets.
The characteristics 1 and 2 above are coming from our previous model [1], whereas points 3 and 4 are the main contributions of the work presented here.
Related works
Multimodal learning with bio-medical data
Several studies used multimodal data in the bio-medical field, but only a few used both imaging and clinical data, as most are focused on using images from different acquisition methods such as PET and MRI. In our case, we deal with both imaging and clinical data, i.e. data having specific dimensions and characteristics for each modality. Among the possible data fusion strategies [2], Xu et al. inspired us with an intermediate-fusion-based neural network. The images go through convolution layers, while the clinical data go through fully-connected layers. Eventually, the two sub-networks are fused into a fully-connected neural network for classification.
Bhagwat et al. [3] employed Siamese Networks to use data (Region Of Interest (ROI) measures from MRI, and cognitive scores) to predict cognitive decline from two medical visits. Siamese neural networks are used to perform comparisons of pairs of data. They are made of two identical branches (same layers and same parameters) which share weights from initialization to the end of training and are fed with paired inputs. Siamese networks have shown great performance for different applications such as handwriting recognition [4] or facial recognition [5]. Finally, Lee et al. [6] used a multimodal recurrent neural network to detect subjects converting to Alzheimer’s disease. Their approach is interesting but lacks a joint learning part and, like Bhagwat’s, relies on the computation of ROI-based metrics, which can be imprecise and lead to detection errors.
In order to circumvent the shortcomings of the methods above, we have proposed the Multimodal3DSiameseNet model [1] in 2020, which is able to detect the cognitive decline of Alzheimer’s patients using multimodal data (clinical data and 3D MRI). In [1], we showed that for Alzheimer’s disease, our architecture, when using only two medical visits, gives better predictions for cognitive decline than Recurrent Neural Network models using either three or four successive visits. However, our training protocol was based on the use of two visits for each patient, with strong constraints on their times (\(T_0\) + 6 months or \(T_0\) + 12 months).
Transfer learning strategy
The transfer learning approach consists in using a model already trained on a large quantity of data that presents similarities to the data of interest. This model’s architecture and its parameters are then used as a starting point for a second training, with a smaller training set, for the final application. The main appeal of this strategy is that it is supposed to help the network converge quicker than using a random initialization [7]. We need to choose which layers (or sub-modules) will be re-trained and which ones will have their parameters frozen (i.e. not updated during re-training). In the medical imaging field, transfer learning can be done by using images from a study on the same disease but from another population or another imaging modality, or from a different disease on the same organ. A recent example is the transfer from lung images obtained with X-rays to scanner images for the diagnosis of COVID-19 [8].
Parkinson dedicated studies
In this section, we focus on recent research about Parkinson’s disease, which is a very active field. Focusing only on clinical data, some large studies show the importance of long-term cognitive trajectories on a large number of patients [9]. The authors in [9] have focused on Mini-Mental State Examination with classical machine learning methods to extract long-term predictions. Different studies [10, 11] have used specific tools to extract the cerebellar and subcortical features from MRI, before applying some classical machine learning algorithms to obtain multiple indicators to assist the clinical diagnosis.
Other approaches used deep learning models [12] to extract deep features from PET images to classify different Parkinson’s disease states (early idiopathic Parkinson’s disease and atypical parkinsonian syndromes). Chandaran et al. [13] have proposed a recent review on transfer learning usage for different brain diseases using MRI. They show great possibilities of transfer learning approaches, however, they also mentioned that it is not always very accurate. Also using a transfer learning approach, and concerning Parkinson’s disease, Basnin et al. [14] have proposed a model using a pre-trained DenseNet architecture to extract deep features from MRI and an LSTM model to discover temporal dependencies. Their study is not multimodal and is focused only on MRI.
Method
Multimodal deep learning model architecture: multimodal3DSiameseNet
This work aims to propose an adaptable deep neural network architecture designed to make long-term prognosis on the evolution of neurological diseases in subjects, to identify those that are more at risk. In our previous work [1], we proposed a model for detecting the cognitive decline of Alzheimer’s patients based on two visits, consisting necessarily of the baseline visit and either the 6-month or 12-month follow-up visit.
In the present work, we re-use this model with slight changes for new tasks:
-
long-term (up to 72-month) prediction of the cognitive decline of Alzheimer’s patients, from two early visits \(T_i\) and \(T_i+\delta \) picked randomly (between 0 and 24 months) among the available visits for each patient between the baseline and 24 months, with no specific fixed times for the two visits considered by the model, nor fixed time interval between these two visits;
-
transfer learning from one neurodegenerative disease (Alzheimer’s) to another neurodegenerative disease (Parkinson’s)
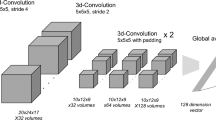
Figure 1 shows the general architecture of our model: Multimodal3DSiameseNet. To use multimodal data, our model is divided into three sub-modules, two of them being siamese networks, and the third one is a fully connected network for risk factors common to all patients. This allows us to obtain different points of view on the disease’s evolution: a morphological point of view with the medical images, a cognitive point of view with the clinical tests, and complementary information coming from risk factors.
Compared to the model we introduced in [1], small changes have been made in the 3D SiameseNet part (explained in the next subsection). The training protocol has also been modified so that the output classes are either “stable” or “declining”, and correspond to a long-term prediction regarding the progression of the disease (up to 72 months), inferred from two medical visits picked randomly amongst the subject’s available visits. We shared our code on a specific GitHub page.Footnote 1
Updated version of our Multimodal3DSiameseNet model, derived from [1]
3D convolutional neural network for MR images
In the first version of this model [1], we used average pooling layers after each convolution, but in the model we use in this paper, pooling is replaced by convolutions with a 3\(\times \)3\(\times \)3 pixels kernel and a stride of two pixels in the three dimensions to be able to optimize the dimension reduction during training. Indeed, average pooling layers lose more information than convolutional 3D layers with stride 2 as authors mentioned in [15].
The two branches of this siamese neural network are then combined by computing the absolute difference between the output features of each branch. This allows us to extract a high-level representation of the morphological changes between any two pairs of visits at \(T_i\) and \(T_i+\delta \). Then this result is flattened to obtain a 1D vector that will be fused with the features obtained from clinical data.
Feed forward networks for clinical data
Qualitative and quantitative clinical data are processed at the same time as the medical images. Clinical scores from visits \(T_i\) and \(T_i+\delta \) will be fed to a second siamese network, made of two feed-forward branches. These two branches are combined using the absolute difference operation. The risk factors (age at visit \(T_i\), gender, and genomics) are processed independently with a simple feed-forward network. These two types of clinical information are then concatenated into a single feature vector representing the clinical evolution of the patient between the two visits.
Modality fusion
Finally, the features from the different modalities are merged using an intermediate fusion strategy: first they are concatenated, then fed to a succession of fully connected layers. These joint layers allow the network to leverage correlation between the modalities.
Datasets
Datasets construction
Data used to pre-train the model come from the public database Alzheimer’s Disease Neuroimaging Initiative (ADNI) [16]. It is based on a multicentric study on Alzheimer’s disease development in the Elderly and is a good use case for our model as the cohort is relatively large (800 subjects in the original cohort ADNI-1), all subjects undergo a medical visit every 6 months for up to 72 months, and multiple modalities of data are available (at least for up to 24 months in the earlier version of the dataset): MRI, PET, genetics, demographics, biological measures, and cognitive scores. Subjects in this study receive a professional diagnosis at each 6-monthly visit: healthy (i.e. normal control (NC)), mild cognitive impairment (MCI), or Alzheimer’s disease (AD). The ADNI-1 cohort is composed of 200 NC, 400 MCI, and 200 AD subjects (based on the diagnosis at inclusion). These diagnosis give us information about the health of the subjects, but as they are punctual, they do not give insights into the long-term evolution of the pathology.
The clinical variables available in the ADNI database are demographic data (age, gender), genomics (APOE4 alleles), and eight more clinical attributes (results of cognitive tests, and biological measures). More details are given in “Training the model with ADNI data” section.
In this study, we use all subjects from the ADNI-1 cohort who had MR images taken at baseline, 6 months, 12 months, and 24 months, obtaining 381 subjects in total. For each subject, we form six pairs that correspond to the possible combinations of two available visits: {baseline, 6 months}, {6 months, 12 months}, {12 months, 24 months}, {baseline, 12 months}, {baseline, 24 months}, and {6 months, 24 months}.
For the transfer learning stage, we use data from the Parkinson’s Progression Markers Initiative (PPMI) database [17]. This database is a longitudinal study of 200 control patients and 400 patients with Parkinson’s disease. They have a medical visit at inclusion and after 12 months, 24 months, and 36 months. Similarly to the ADNI database, we consider structural brain MRI as well as cognitive tests and information about risk factors. As these modalities are similar as ADNI’s modalities, the PPMI dataset is a good candidate for our transfer learning experiment from Alzheimer’s disease to Parkinson’s disease. We only use the pairs {baseline, 12 months} for fine-tuning, giving us 134 pairs (47 stable subjects and 87 declining subjects). The risk factors fed to the model are the same as before, i.e. age, gender, and APOE4 genotype. We use eight of the available clinical scores for the clinical part of the model (see the “Training the model with ADNI data” section for more details).
Ground truth labelling
In our ground-truth, we want to have labels that reflect the long-term cognitive evolution of each patient. But, there is no such information per se in the ADNI dataset. Therefore, we had to create a specific ground truth. For that purpose, we use the same method as Bhagwat et al. [3] to create clusters of subjects based on the long-term evolution of the cognitive score Mini Mental State Evaluation (MMSE) over a 72-months time span. This score is widely used for the assessment of Alzheimer’s disease progression, which makes it a good marker for cognitive decline, and varies between 30 (best score, healthy patient) and 0. This clustering reveals two groups: a “Stable” group in which the MMSE results stay high over time, and a “Declining” group in which the MMSE results drop over time [3].
Using these results, every subject is assigned to either the Stable or Declining cluster. These clusters are linked to the diagnosis given with the ADNI dataset but do not bear the same information, as shown in Table 1. Indeed, while all normal control (NC) patients are stable and most Alzheimer’s disease (AD) are unsurprisingly classified as experiencing a long-term decline, 45% of the mild cognitive impairment (MCI) population is clustered as Stable, and 55% as Declining.
Finally, we obtain after preprocessing and labeling a total of 2268 pairs of visits, divided into 1197 Stable pairs (= negative pairs), and 1071 Declining pairs (= positive pairs). It is important to note that, because we chose the MMSE score to be our reference for ground truth assignment, this score will not be a part of the clinical data that we will feed to our model.
For the PPMI data, we similarly created a long-term ground truth to the ADNI data, using the Unified Parkinson Disease Rating Scale (UPDRS) score for the clustering, as a disease progression score (similar to MMSE for Alzheimer’s). The subjects were grouped according to their cognitive decline or stability over a 36 months time span. Similarly to the ADNI subjects, we have a group of stable subjects and a group of declining subjects whose UPDRS scores increase with time. Similarly to ADNI, we exclude the UPDRS score from the input data.
MRI and clinical variables pre-processing and augmentation
To remove unnecessary information in the images from both ADNI and PPMI data, the brain MR images have been preprocessed with alignment and skull-stripping as in [18], then cropped to remove most of the background. Finally, because the subjects’ brains have different sizes, we re-sized all of the cropped images to 204\(\times \)216\(\times \)150 pixels then downsized them to 102\(\times \)108\(\times \)75 pixels.
Let us now focus on attribute selection. For ADNI clinical data, we start by removing the variables with a large number of missing values (mostly bio-markers). Fortunately, as we showed in [1], using MRI in our model partly compensates for these missing clinical values. Then, based on pairwise Spearman correlations between variables, we select one variable for each group of highly correlated variables (excluding the MMSE scores). In total, we select the three risk factors AGE, GENDER, and APOE4 alleles, and eight cognitive scores: LDELTOTAL, RAVLT learning, RAVLT immediate, CDRSB, FAQ, TRABSCOR, RAVLT forgetting, and DIGITSCOR.
For PPMI data, using a similar procedure, we select the same risk factors, and eight of the 13 available clinical scores, namely HY, NHY, UPSIT, HVLT, LNS, QUIP, SCOPA, STAI. The score UPDRS, available in PPMI data, is the disease progression score used that we use for creating our ground-truth (so we do not use it as an input clinical variable).
As it is commonly the case, to improve training and reduce risks of overfitting, we use on-the-fly image augmentation during training, to introduce variability in the brain images. To do this, we randomly apply a mix of Gaussian blur, rotations, flips, and contrast modifications.
Experiments and results
Training the model with ADNI data
For pre-training with the ADNI dataset, data is split at the subject level: we take 60% for training (229 subjects), 20% for validation (76 subjects), and 20% for testing (76 subjects). For each subject, 4 visits are most often available, producing 6 possible pairs of visits for each. The total number of pairs for the test set is 456 but, as it misses some visits, we only have 450 pairs of visits in our test dataset. To avoid bias during the evaluation of the model, all the visit pairs from a single subject belong to only one subset of data (training, validation, or test). We trained our multimodal model with a four-fold stratified cross-validation protocol between training and validation sets. The test set (76 patients) is identical for all folds.
Results and influence of short-term variations
After training the models, we obtained a mean accuracy of 0.91 on our ADNI test dataset, with a standard deviation of 0.01, and a mean F1 score of 0.90 with a standard deviation of 0.01 over our four models (one per cross-validation fold). The histograms in Fig. 2 show the distribution AD, MCI, and NC diagnosis, at the time of the second visit for each pair, among the True Positives (patients correctly predicted by our model as declining), the True Negatives (patients correctly predicted as stable), the False Positives and the False Negatives.
Distribution of Control (NC), Mild Cognitive Impairment (MCI), and Dementia (AD) diagnosis among the True Positives (TP), the True Negatives (TN), the False Positives (FP) and the False Negatives (FN). Values on top of each column are the average counts over the 4 cross-validated models: for instance, on average over the 4 models (450 visit pairs in the test dataset), 187 visit pairs lead to a True Positive prediction (“Declining” prediction, in the presence of a decline in the ground-truth). In blue in the fourth column, we can see that more than 75% of them were diagnosed as having dementia (AD) during the second visit we used as input
As expected, the NC and AD subjects are mostly correctly classified into the Stable and Declining classes. This confirms that both subjects with no pathological signs (Alzheimer’s disease or other) and severely affected subjects are effectively detected by our model as (respectively) Stable and Declining. But, on average on the 4 folds, more than 28 patients with Mild Cognitive Impairment (MCI) are misclassified (8 as False Positives, and 20 as False Negatives). Since MCI subjects form a heterogeneous group (MCI subjects can be either stable or declining, as shown in Table 1), we need to look more closely at their MMSE scores to interpret these classification errors.
Most of these errors probably come from the fact that we train and make inferences with our model only using the evolution of the patients from 2 visits (randomly chosen between the baseline and 24 months), whereas we created our ground-truth using the long-term evolution of the MMSE score (over a 72-month time span).
So, in Fig. 3, we compare, for each patient, the MMSE value from the latest visit of each pair used by our model for prediction with the MMSE used to create the ground-truth (last MMSE value available for each subject, up to 72 months after inclusion).
Top: Distribution of the last available MMSE values (used for creating the ground-truth, up to 72 months depending on the patient) among TN, FP, FN, and TP. Bottom: Distribution of the second visit’s MMSE (for pairs of visits used by the model, corresponding to intervals between 6 and 24 months). The white color corresponds to a MMSE of 26, which is the threshold for Alzheimer’s disease [19]. Values are the average counts over the 4 cross-validation folds
From this figure, we can see that, obviously, these two MMSE scores are mostly high for TN patients and low for TP patients.
This figure also shows that a majority of false negatives seem to correspond to pairs where the MMSE value is low at the end of the study (used for ground-truth), but still high at the second visit that we used for prediction. We can interpret this as being subjects who developed Alzheimer’s disease after the second visit fed to our model. This means that, although our model has a good overall performance to predict long-term evolution of subjects (F1 score of 0.90), it is inherently limited to detecting the cognitive decline only when it is perceptible in the short-term time frame used for the study. In particular, for subjects who started to decline after the second visit used for inference, our model cannot accurately predict their decline.
On the other hand, studying Fig. 3 does not help with the interpretation of FPs, as the pairwise MMSE distributions are not very different from the long-term MMSE distributions for FPs. However, there are fewer FPs than FNs (on average on the 4 folds, 10.5 v.s. 32), and this might be an artifact due to the way we create our ground-truth (using automatic clustering, without supervision from a medical doctor). For example, if these patients have a punctual drop in their MMSE score on their last visit, they will be considered in our ground-truth as declining (Positive), even though this might just be that they were not in a good shape on the day of their last visit.
Influence of time interval
Following the findings from the above section, we decided to have a closer look at the influence of the time interval \(\delta \) between two visits used by our model, on the model’s prediction performance. For example, we expected the pairs “baseline and 24 months” to be the best-classified pairs, given that it corresponds to the longest possible interval in the training and validation datasets.
But, Table 2 shows that on average and contrary to what we expected, increasing \(\delta \) does not improve the classification results. Indeed, all pairwise T-tests for different time intervals give p-values between 0.084 and 0.35 (and preliminary pairwise Fisher test p-values are all very high). This means that our model is at least partly invariant with the time interval between the two input visits.
In a medical longitudinal study, the independence of prognosis performance to time intervals is an interesting and useful characteristic of our model because it means that we do not necessarily need a patient’s follow-up over a long time span to predict its long-term evolution.
Fine-tuning results with PPMI data
For transfer learning from Alzheimer’s disease (ADNI dataset) to Parkinson’s disease (PPMI dataset), we tried and compared several initialization and re-training strategies:
-
(1) Training the whole network from scratch (i.e. with randomly initialized parameters)
-
(2) Fine-tuning all the layers
-
(3) Fine-tuning the MRI and joint sub-modules and training the clinical sub-module from scratch
-
(4) Freezing all convolution layers, fine-tuning the fully-connected layers of the MRI and joint sub-modules and training the clinical sub-module from scratch
These four models were trained with three-fold cross-validation and the same fold distribution. Figure 4 shows the evolution of the average validation loss over successive training epochs. The two following subsections are focusing on the compared performances of our four strategies at training epochs number 5 and 30 respectively, to compare the convergence speeds of our four strategies.
At epoch 5
Our experimental comparison of the 4 strategies above, once the 5th training epoch has just been completed, is illustrated in Table 3 and Fig. 5.
At epoch 5, the model using strategy (1) (whole model trained from scratch on PPMI) did not yet learn anything, as the value of the validation loss never decreased.
The three remaining models (2)–(4) all have a low true positives to false positives ratio, but strategy (4) seems to be the most effective at an early stage of learning for Parkinson subjects’ prediction of cognitive decline, as shown in the ROC curves in Fig. 5.
At epoch 30
As expected, once the models have been trained for 30 epochs, most performance metrics for all models have improved, except for the test accuracy of model (3) and the F1 measure of model (2). The AUC measures, which in our view are the most important ones, have all increased (see Table 4 and Fig. 6). The sub-par performance of model (2) can be obviously explained by the fact that it used pre-trained weights for the clinical part of the network, while the clinical scores are not the same between ADNI and PPMI.
Models (3) and (4) are better than model (1) trained from scratch on PPMI data. This shows that transfer learning improves the prediction accuracy compared to “classical” learning (from scratch), at least on a small target dataset such as the PPMI subset considered here. We note that the difference between the performances of strategies (3) and (4) is not statistically significant, as showed by a Kruskal–Wallis test on the F1-scores (p-value = 0.08798 > 0.05).
Discussion
Our experiments with the improved Multimodal3DSiamese Net model on the ADNI data showed that its long-term prediction accuracy for the cognitive decline does not vary much with the time interval between the two input visits. This is an important result, as it would normally be expected that a longer interval leads to a better prediction. For example, in [20], the authors conceived a model to evaluate the probabilities of transition from healthy to mild cognitive impairment to Alzheimer’s disease and found that the transition probability increased with the time interval between visits. In comparison, thanks to our model’s architecture and our training dataset and protocol, we can use two early medical visits to predict the long-term evolution of a patient, and these two visits can be chosen randomly among the early visits available for each patient. Of course, this strategy will only work if the subject shows signs of cognitive decline in the time frame considered by our model.
However, we also found out that some of our model’s prediction errors were probably caused by punctual variations in the score (MMSE) used to label our classes of subjects (the score from each subject’s last visit). This may mean that our choice of ground truth (i.e. automatic generation using clustering), even if it is quite commonly used in the community, is not as accurate as it could be. Our work would certainly benefit from an Alzheimer’s disease expert’s opinion on the ground-truth that we use.
Finally, our transfer learning experiments on the small subset of PPMI dataset on Parkinson’s disease showed a quicker optimization and better classification performance when transferring the knowledge from another neurodegenerative disease (here, Alzheimer’s), compared to training from scratch.
We are mainly interested in strategies (3) and (4), which are based on transfer learning on only the modalities which are similar in the source and target datasets (here MRIs and risk factors), with the difference that method (4) does not fine-tune the convolutional layers (it freezes them). Among these two strategies, we have shown that there is no significant difference in their performances. In terms of computational complexity, the most interesting one is model (4) with about 2 million parameters to learn, whereas model (3) has more than 35 million parameters to train. This is why we would recommend using strategy (4), where we do not need to fine-tune the convolutional layers (provided the two diseases considered for the transfer have similar enough characteristics in terms of imagery). In particular, we showed that we can use, for Parkinson’s patients, the 3D filters learned from Alzheimer’s, hence saving a great amount of computational effort. This paves the way to the possibility of re-using, easily and with very limited computational cost, our pre-trained model for the long-term prediction of other neurodegenerative diseases (beyond Parkinson’s), even for rare or under-studied diseases where only a limited dataset is available.
Conclusion
In this work, we focused on predicting the cognitive decline of patients affected by neurodegenerative diseases, based on multimodal data (brain MRIs, clinical tests, and various risk factors). The model that we propose to use, Multimodal3DSiameseNet, is able to process directly the 3D MRI. We showed that this model gives a good prognosis for Alzheimer’s disease progression in the long term (up to 72 months), even when using as inputs two early medical visits picked randomly among the available early visits. Contrary to what we initially expected, the time interval between the two visits considered for a given subject does not seem to affect greatly the accuracy of the prediction for that patient.
This is an interesting characteristic of our model and experimental protocol, especially when dealing with neurodegenerative diseases, where it is quite frequent that the patients miss some control visits. However, we found out that, in the future, we need to discuss our ground-truth with an expert on Alzheimer’s disease, as it has been created automatically and, in some cases, it might be biased by the punctual mental state of the patients during their last visit. We are also currently collaborating with an Alzheimer’s disease expert so as to analyze more in-depth the differences in predictions depending on the time interval between the two visits given as input to the system.
We also showed the adaptability of our Multimodal3D SiameseNet model to different diseases, with the example of transfer learning from Alzheimer’s to Parkinson’s. Based on the comparison of different transfer learning strategies, we showed that, provided we have similar modalities in both datasets, we can easily, and with limited computational complexity, transfer the knowledge learned by our model from a given neurodegenerative brain disease to another.
As Deep Learning can achieve impressive effectiveness, but at the cost of big annotated datasets, this is a very interesting feature of our model and protocol. In particular, our adaptable multimodal model could be used for long-term prediction of cognitive decline in a variety of neurodegenerative diseases which are too rare or under-studied to have large dedicated labelled datasets.
Beyond neurodegenerative diseases, in the future, we plan to transfer our pre-trained model to other brain-related disorders which involve a temporal evolution, including for instance the detection of relapse risk after drug withdrawal.
References
Ostertag C, Beurton-Aimar M, Visani M, Urruty T, Bertet K (2020) Predicting brain degeneration with a multimodal siamese neural network. In: 2020 tenth international conference on image processing theory, tools and applications (IPTA). IEEE, pp 1–6
Ramachandram D, Taylor GW (2017) Deep multimodal learning: a survey on recent advances and trends. IEEE Signal Process Mag 34(6):96–108
Bhagwat N, Viviano JD, Voineskos AN, Chakravarty MM (2018) Modeling and prediction of clinical symptom trajectories in Alzheimer’s disease using longitudinal data. PLoS Comput Biol 14(9):1006376
Bromley J, Guyon I, LeCun Y, Säckinger E, Shah R (1994) Signature verification using a “siamese” time delay neural network. In: Advances in neural information processing systems, pp 737–744
Lin S, Zhao Z, Su F (2016) Homemade ts-net for automatic face recognition. In: Proceedings of the 2016 ACM on international conference on multimedia retrieval, pp 135–142
Lee G, Nho K, Kang B, Sohn K-A, Kim D (2019) Predicting Alzheimer’s disease progression using multi-modal deep learning approach. Sci Rep 9(1):1–12
Donahue J, Jia Y, Vinyals O, Hoffman J, Zhang N, Tzeng E, Darrell T (2014) Decaf: a deep convolutional activation feature for generic visual recognition. In: International conference on machine learning, pp 647–655
Niu S, Liu M, Liu Y, Wang J, Song H (2021) Distant domain transfer learning for medical imaging. IEEE J Biomed Health Inform 25(10):3784–3793
Wu Y, Jia M, Xiang C, Lin S, Jiang Z, Fang Y (2022) Predicting the long-term cognitive trajectories using machine learning approaches: a Chinese nationwide longitudinal database. Psych Res 310:114434
Ya Y, Ji L, Jia Y, Zou N, Jiang Z, Yin H, Mao C, Luo W, Wang E, Fan G (2022) Machine learning models for diagnosis of Parkinson’s disease using multiple structural magnetic resonance imaging features. Front Aging Neurosci 14:808520
Shibata H, Uchida Y, Inui S, Kan H, Sakurai K, Oishi N, Ueki Y, Oishi K, Matsukawa N (2022) Machine learning trained with quantitative susceptibility mapping to detect mild cognitive impairment in Parkinson’s disease. Parkinsonism Relat Disorders 94:104–110
Wu P, Zhao Y, Wu J, Brendel M, Lu J, Ge J, Bernhardt A, Li L, Alberts I, Katzdobler S, Yakushev I, Hong J, Xu Q, Sun Y, Liu F, Levin J, Höglinger G, Bassetti C, Guan Y, Oertel WH, Weber WA, Rominger A, Wang J, Zuo C, Shi K (2022) Differential diagnosis of parkinsonism based on deep metabolic imaging indices. J Nucl Med 64(1):1741–1747
Chandaran SR, Muthusamy G, Sevalaiappan LR, Senthilkumaran N (2022) Deep learning-based transfer learning model in diagnosis of diseases with brain magnetic resonance imaging. Acta Polytech Hung 19:179
Basnin N, Nahar N, Anika FA, Hossain MS, Andersson K (2021) Deep learning approach to classify Parkinson’s disease from MRI samples. In: International conference on brain informatics, pp 536–547
Springenberg JT, Dosovitskiy A, Brox T, Riedmiller MA (2015) Striving for simplicity: the all convolutional net. In: International conference on learning representations (ICLR) workshops . http://arxiv.org/abs/1412.6806
Jack CR Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, Whitwell L, Ward JC (2008) The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imag 27(4):685–691
Marek K, Jennings D, Lasch S, Siderowf A, Tanner C, Simuni T, Coffey C, Kieburtz K, Flagg E, Chowdhury S (2011) The parkinson progression marker initiative (PPMI). Prog Neurobiol 95(4):629–635
Korolev S, Safiullin A, Belyaev M, Dodonova Y (2017) Residual and plain convolutional neural networks for 3D brain MRI classification. In: International symposium on biomedical imaging (ISBI), pp 835–838
Perneczky R, Wagenpfeil S, Komossa K, Grimmer T, Diehl J, Kurz A (2006) Mapping scores onto stages: mini-mental state examination and clinical dementia rating. Am J Geriatr Psychiatr 14(2):139–144
Evans S, McRae-McKee K, Hadjichrysanthou C, Wong MM, Ames D, Lopez O, de Wolf F, Anderson RM (2019) Alzheimer’s disease progression and risk factors: a standardized comparison between six large data sets. Alzheimer’s Dementia Transl Res Clin Interv 5(1):515–523
Acknowledgements
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp- content/uploads/how_to_apply/ ADNI_Acknowledgement_ List.pdf. One of the goals of ADNI has been to test whether magnetic resonance imaging(MRI) and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). For up-to-date information, see www.adni-info.org. Data used in the preparation of this article were obtained from the Parkinson’s Progression Markers Initiative (PPMI) database (www.ppmi-info.org/data). For up-to-date information on the study, visit www.ppmi-info.org. PPMI—a public-private partnership—is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners.
Funding
This study was funded by the French region “Nouvelle-Aquitaine” (Grant Number RCB 2018-A02699-46).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
the authors declare that they have no conflict of interest.
Consent to participate
This article does not contain patient data.
Consent for publication
All authors have checked the manuscript and have agreed to the submission.
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
For the Alzheimer’s Disease Neuroimaging Initiative (see “Acknowledgements”).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ostertag, C., Visani, M., Urruty, T. et al. Long-term cognitive decline prediction based on multi-modal data using Multimodal3DSiameseNet: transfer learning from Alzheimer’s disease to Parkinson’s disease. Int J CARS 18, 809–818 (2023). https://doi.org/10.1007/s11548-023-02866-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-023-02866-6