Abstract
Purpose
The accuracy improvement in endoscopic image classification matters to the endoscopists in diagnosing and choosing suitable treatment for patients. Existing CNN-based methods for endoscopic image classification tend to use the deepest abstract features without considering the contribution of low-level features, while the latter is of great significance in the actual diagnosis of intestinal diseases.
Methods
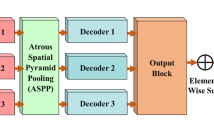
To make full use of both high-level and low-level features, we propose a novel two-stream network for endoscopic image classification. Specifically, the backbone stream is utilized to extract high-level features. In the fusion stream, low-level features are generated by a bottom-up multi-scale gradual integration (BMGI) method, and the input of BMGI is refined by top-down attention learning modules. Besides, a novel correction loss is proposed to clarify the relationship between high-level and low-level features.
Results
Experiments on the KVASIR dataset demonstrate that the proposed framework can obtain an overall classification accuracy of 97.33% with Kappa coefficient of 95.25%. Compared to the existing models, the two evaluation indicators have increased by 2% and 2.25%, respectively, at least.
Conclusion
In this study, we proposed a two-stream network that fuses the high-level and low-level features for endoscopic image classification. The experiment results show that the high-to-low-level feature can better represent the endoscopic image and enable our model to outperform several state-of-the-art classification approaches. In addition, the proposed correction loss could regularize the consistency between backbone stream and fusion stream. Thus, the fused feature can reduce the intra-class distances and make accurate label prediction.






Similar content being viewed by others
References
Favoriti P, Carbone G, Greco M, Pirozzi F, Pirozzi REM, Corcione F (2016) Worldwide burden of colorectal cancer: a review. Updat Surg 68(1):7–11
Korbar B, Olofson AM, Miraflor AP, Nicka KM, Suriawinata MA Torresani L, Suriawinata AA, Hassanpour S (2017) Deep-learning for classifification of colorectal polyps on whole-slide images. J Pathol Inf 8(1):1–24
Jia X, Xing X, Yuan Y, Xing L, Meng QH (2019) Wireless capsule endoscopy: a new tool for cancer screening in the colon with deep-learning-based polyp recognition. Proc IEEE PP(99):1–20
Karkanis SA, Iakovidis DK, Maroulis DE, Karras DA, Tzivras M (2003) Computer-aided tumor detection in endoscopic video using color wavelet features. IEEE Trans Inf Technol Biomed A Publ IEEE Eng Med Biol Soc 7(3):141
Alexandre LA, Nobre N, Casteleiro J (2008) Color and position versus texture features for endoscopic polyp detection. In: International conference on biomedical engineering & informatics
Yang J, Chang L, Li S et al (2020) WCE polyp detection based on novel feature descriptor with normalized variance locality-constrained linear coding. Int J Comput Assist Radiol Surg 15(8):1291–1302
Bi L, Feng DDF, Fulham MJ, Kim J (2020) Multi-label classification of multi-modality skin lesion via hyper-connected convolutional neural network. Pattern Recogn 107:107502
Ding X, Zhang X, Ma N, Han J, Ding G, Sun J (2021) Repvgg: making VGG-style convnets great again. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 13 733–13 742
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In IEEE conference on computer vision and pattern recognition (CVPR) 2016, pp 770–778
Xie S, Girshick R, Dollár P, Tu Z, He K (2017) Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1492–1500
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Shin Y, Balasingham I (2017) Comparison of hand-craft feature based SVM and CNN based deep learning framework for automatic polyp classification. In: Engineering in medicine & biology society
Guo X, Yuan Y (2019) Triple ANET: adaptive abnormal-aware attention network for WCE image classification. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 293–301
Xing X, Yuan Y, Meng QH (2020) Zoom in lesions for better diagnosis: attention guided deformation network for WCE image classification. IEEE Trans Med Imaging PP(99):1
Guo X, Yuan Y (2020) Semi-supervised WCE image classification with adaptive aggregated attention. Med Image Anal 64:101733
Poudel S, Kim YJ, Vo DM, Lee SW (2020) Colorectal disease classification using efficiently scaled dilation in convolutional neural network. IEEE Access PP(99):1
Schlemper J, Oktay O, Schaap M, Heinrich M, Kainz B, Glocker B, Rueckert D (2019) Attention gated networks: learning to leverage salient regions in medical images. Med Image Anal 53:197–207
Zhang J, Xie Y, Xia Y et al (2019) Attention residual learning for skin lesion classification. IEEE Trans Med Imaging 38(9):2092–2103
Woo S, Park J, Lee J-Y, Kweon IS (2018) Cbam: convolutional block attention module. In: Proceedings of the European conference on computer vision (ECCV), pp 3–19
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241
Jia X, Mai X, Cui Y, Yuan Y, Xing X, Seo H, Xing L, Meng MQ (2020) Automatic polyp recognition in colonoscopy images using deep learning and two-stage pyramidal feature prediction. IEEE Trans Autom Sci Eng 17(3):1570–1584
Lin TY, Dollar P, Girshick R, He K, Hariharan B, Belongie S (2017) Feature pyramid networks for object detection. In: 2017 IEEE conference on computer vision and pattern recognition (CVPR)
Kong T, Yao A, Chen Y, Sun F (2016) Hypernet: towards accurate region proposal generation and joint object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 845–853
Li X, Wang W, Hu X, Yang J (2019) Selective kernel networks. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 510–519
Pogorelov K, Randel KR, Griwodz C, Eskeland SL, de Lange T, Johansen D, Spampinato C, Dang-Nguyen D-T, Lux M, Schmidt PT et al. (2017) Kvasir: a multi-class image dataset for computer aided gastrointestinal disease detection. In: Proceedings of the 8th ACM on multimedia systems conference, pp 164–169
Alaskar H, Hussain A, Al-Aseem N et al (2019) Application of convolutional neural networks for automated ulcer detection in wireless capsule endoscopy images. Sensors 19(6):1265
Sun G, Cholakkal H, Khan S, Khan F, Shao L (2020) Fine-grained recognition: accounting for subtle differences between similar classes. In: Proceedings of the AAAI conference on artificial intelligence, vol 34, no 7, pp 12047–12054
Klare P, Sander C, Prinzen M, Haller B, Nowack S, Abdelhafez M, Poszler A, Brown H, Wilhelm D, Schmid RM et al (2019) Automated polyp detection in the colorectum: a prospective study (with videos). Gastrointest Endosc 89(3):576–582
Wang P, Berzin TM, Brown JRG, Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D et al (2019) Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut 68(10):1813–1819
Jorge Bernal, Fernando Vilarino, Gloria Fernandez-Esparrach, Debora Gil, Sanchez JF (2015) WM-DOVA maps for accurate polyp highlighting in colonoscopy: validation vs. saliency maps from physicians. Comput Med Imaging Graph 43:99–111
Acknowledgements
This work was supported by National Science Foundation of P.R. China (Grants: 61873239), Key R& D Program Projects in Zhejiang Province (Grant: 2020C03074).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, S., Yao, J., Cao, J. et al. Effective high-to-low-level feature aggregation network for endoscopic image classification. Int J CARS 17, 1225–1233 (2022). https://doi.org/10.1007/s11548-022-02591-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-022-02591-6