Abstract
Purpose
COVID-19 has spread rapidly worldwide since its initial appearance, creating the need for faster diagnostic methods and tools. Due to the high rate of false-negative RT-PCR tests, the role of chest CT examination has been investigated as an auxiliary procedure. The main goal of this work is to establish a well-defined strategy for 3D segmentation of the airways and lungs of COVID-19 positive patients from CT scans, including detected abnormalities. Their identification and the volumetric quantification could allow an easier classification in terms of gravity, extent and progression of the infection. Moreover, these 3D reconstructions can provide a high-impact tool to enhance awareness of the severity of COVID-19 pneumonia.
Methods
Segmentation process was performed utilizing a proprietary software, starting from six different stacks of chest CT images of subjects with and without COVID-19. In this context, a comparison between manual and automatic segmentation methods of the respiratory system was conducted, to assess the potential value of both techniques, in terms of time consumption, required anatomical knowledge and branch detection, in healthy and pathological conditions.
Results
High-quality 3D models were obtained. They can be utilized to assess the impact of the pathology, by volumetrically quantifying the extension of the affected areas. Indeed, based on the obtained reconstructions, an attempted classification for each patient in terms of the severity of the COVID-19 infection has been outlined.
Conclusions
Automatic algorithms allowed for a substantial reduction in segmentation time. However, a great effort was required for the manual identification of COVID-19 CT manifestations. The developed automated procedure succeeded in obtaining sufficiently accurate models of the airways and the lungs of both healthy patients and subjects with confirmed COVID-19, in a reasonable time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coronavirus disease 2019 (COVID-19) is a highly infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The first human cases of COVID-19 were recognized in Wuhan, Hubei, China, in late December 2019. By metagenomic RNA sequencing and virus isolation from throat swab samples, a novel coronavirus was identified as the causative agent of this emerging disease [1]. SARS-CoV-2 is a beta coronavirus that is constituted by a single-stranded RNA structure belonging to the Coronaviridae family [2]. Typical features of COVID-19 include endothelial barrier disruption, dysfunctional alveolar-capillary oxygen transmission, reduced oxygen diffusion capacity, alveolar wall thickening, increased vascular permeability and pulmonary oedema [3].
The reference standard in COVID-19 diagnosis is the reverse transcription polymerase chain reaction (RT-PCR). However, the sensitivity of RT-PCR tests is not perfect, with the possibility of false-negative test results [4]. Several reports have indicated a possible role for chest computed tomography (CT) examination in presumptive diagnosis in patients with a high suspicion of infections of SARS-CoV-2. CT imaging may also be helpful in monitoring disease evolution and evaluating therapeutic efficacy [4,5,6].
Utilizing CT images, it is possible to obtain 3D models, by means of segmentation, to allow the assessment of the extent and the distribution of the pathology, providing a useful tool for clinicians. Moreover, a volumetric quantification of the affected regions of the lung lobes can be performed [7]. Actually, both CT and magnetic resonance imaging (MRI) technologies could be adopted for the imaging and measurement of airways; however, CT is the established modality in the field [8].
The medical and bioengineering literature refers to different issues relating to image segmentation for the study of the respiratory system. Besides the quality of scans themselves (e.g. resolution and slice thickness), the appearance of airways on CT scans depends on their diameter and the orientation with respect to the plane of the slice [9]. The largest airways are easier to detect because of their dimensions, the thickness of their walls and the more homogeneous grayscale values [9]. Due to the size of the voxel, thin or stenosed airways can appear broken or discontinuous, resulting in both under- and over-segmentation errors [10]. Moreover, if the airways appear thin or disrupted, the segmentation can erroneously leak, due to the similar intensity and texture of the surroundings [11]. These issues, together with other image artefacts, lead to the impossibility of determining a global threshold in terms of Hounsfield Units (HU) to segment the airways completely. Additionally, the optimal threshold differs for large versus small airways, due to partial volume effects [10].
Airways segmentation can be performed both manually and through semi-automatic methods. Manual segmentation requires the user to delineate the entire structure slice by slice, resulting in a tedious and extremely time-consuming process: a full identification may require several hours of analysis [12]. The variabilities caused by human interaction and time consumption opened the way to automatic methods. Lo et al. [13] presented a review of 15 different techniques for airways segmentation from CT images. Moreover, there are additional studies in which the authors compare their own automatic algorithm with a manual segmentation, taken as a gold standard, resulting in overall detection sensitivity of about 73% for human data [14], through a morphological approach and 69% sensitivity for canine data [15], through a fuzzy logic approach. Focusing on lungs, there are pulmonary disease processes and anatomical conditions, such as incomplete pulmonary fissures, that may alter their appearance [16]. Since the identification of the lungs is based on the large difference in terms of HU between the air and the surrounding tissues, in the case of dense pulmonary or subpleural abnormalities, some areas may not be included in the segmentation [17]; therefore, specialists need to manually edit the results.
Patients suspected of SARS-CoV-2 infection should undergo non-contrast-enhanced chest CT, using a low-radiation-dose protocol, to minimize radiation burden [4]. A number of studies have been published, discussing the most common CT manifestations in patients affected by COVID-19 [18]. Several imaging findings were reported in more than 70% of proven COVID-19 cases, including ground-glass opacities, bilateral abnormalities, lower lobes involvement and vascular enlargement [18]. Other reported chest CT abnormalities with intermediate incidence involve consolidations, linear opacities, septal thickening and/or reticular pattern, crazy-paving pattern, air bronchogram, pleural thickening, halo signs, bronchiectasis, nodules, bronchial wall thickening and reversed halo signs [18]. Less common findings include pleural effusion, lymphadenopathy and pericardial effusion, which typically occur later in the course of the disease [18]. In Table 1, the most common CT imaging features of COVID-19 [6] are described in detail.
The Radiological Society of North America (RSNA) published guidelines to reduce the uncertainty and the variability in reporting imaging findings potentially related to COVID-19 [19], identifying four different categories. It is worth noting that CT imaging manifestations discussed above are non-specific and may overlap with other diseases, including other viral pneumonia [4, 18]. Consequently, the diagnostic value of chest CT imaging for COVID-19 is limited and a negative CT examination result does not exclude COVID-19. The diagnostic accuracy of chest CT depends on reader experience and the adopted criteria. Moreover, due to the intrinsic bidimensional nature of CT findings, their interpretation may be quite tricky, possibly precluding the accurate measurements of dimensions such as area or volume.
Given these constraints, the aims of the study were (1) to implement a method for 3D segmentation from CT scans, thereby demonstrating the visible pathological effects induced by COVID-19; (2) to obtain accurate 3D models that can be utilized to assess the impact of the pathology, by volumetrically quantifying the extension into the affected areas; (3) to assist with the interpretation and communication of the CT findings.
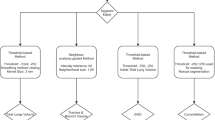
In our study, the most appropriate segmentation techniques were evaluated, beginning with manual segmentation. An automatic procedure was then tested on the same reference patients, to assess its effectiveness. After the identification of the potential value of this automatic method, it was utilized for the segmentation of the respiratory system of COVID-19 positive patients. A conclusive comprehensive comparison, to identify the pros and cons of both the methodologies, in terms of time consumption, required anatomical knowledge, and branch detection sensitivity was conducted.
Materials and methods
The segmentation process was performed using proprietary Mimics software (Materialise, Leuven, Belgium, version 23), starting from different stacks of chest CT images of subjects with and without COVID-19 positive diagnosis. The characteristics of the patients and of the sets of images are listed in Table 2. CT images were downloaded from available online databases [20, 21].
CT images of most common COVID-19 CT findings. Adapted from [6]. a Ground-glass opacity. b Consolidation. c Reticular pattern. d Crazy-paving pattern. e Fibrosis. f Halo sign
The segmentation process of the CT stacks of healthy subjects was carried out manually, to obtain reference segmentation models; then, the automatic Segment Airway tool provided by Mimics Pulmonary Module was applied on them, to verify its efficacy in airway branches identification. In the case of COVID-19 positive patients, just the automatic Segment Airway and Segment Lungs tools were utilized. Even if not strictly important for the visualization or quantification of COVID-19 manifestations, lower airways were anyway always included in the models, both to guarantee a more comprehensive comparison between manual and automatic segmentation tools and to provide a clearer and intuitive anatomical representation.
Healthy subjects segmentation
In the manual segmentation process, a region growing algorithm was used for airway reconstruction, placing a first seed point in correspondence of the trachea. Additional seed points were then placed on the last detected airway branches. This was followed by meticulous manual editing on the single slices. Finally, a global smoothing (Laplacian, I order, with shrinking compensation) and a wrap operation were performed on the converted parts, to enhance the surface finishing and filter small inclusions.
The lungs were segmented first using a thresholding algorithm (threshold − 1024 ÷ − 500 HU); a Boolean operation was used to include just the lung parenchyma, splitting the right and left lung masks. Pulmonary internal vasculature was not included in the segmentation, resulting in masks characterized by vascular cavities; all the isolated vessel branches were then added through combined Boolean operations, to improve the appearance of the final models. The obtained masks were manually edited and then globally smoothed once converted into parts. On the other hand, the automatic Mimics Segment Airway tool allowed to segment the airways starting from two seed points, indicating the start of the trachea and its direction. A middle detection level was set to balance branches identification and leakage occurrence. Global smoothing algorithm was applied as in the previous case. The Mimics Pulmonary Module additionally provides the centreline-labelling tool, which allows an automatic computation of the number of airway branches.
COVID-19 positive patient segmentation
In the automatic segmentation process applied on COVID-19 positive patients, the Segment Airway tool was exploited to obtain airways 3D models and the centreline was extracted as previously explained. The centreline was needed to automatically segment the lungs through the Segment Lungs tool, adopting the default thresholds (− 1024 ÷ − 500 HU). Due to the presence of numerous COVID-19 manifestations, several regions of the lung parenchyma were not included in the masks, because of their high-intensity level. All the masks were refined through a rigorous manual editing operation, to incorporate opacities and correct all the boundaries. Pulmonary vasculature correction and global smoothing were then performed as previously described.
COVID-19 manifestations were identified through accurate visual inspection; a thresholding algorithm was adopted for the segmentation, properly selecting the boundary thresholds case by case and limiting the region of interest (ROI). Afterwards, the obtained masks were filled with one or two voxels levels, to get less sharp contours. The resulting masks were intersected with the masks of the lungs, and then, they were manually adjusted. The final smoothing was limited in this case, to maintain the irregular appearance of the CT findings.
Post-processing
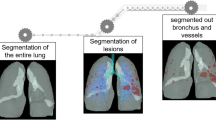
All the parts were later imported in 3-Matic software (Materialise, Leuven, Belgium, version 15), and common operations were carried out on all the models. The airways were divided into seven different portions and hollowed towards the outside independently (Fig. 2), assigning different thickness values according to the literature [22]. The different regions were then merged through a Boolean operation, the connecting edges were smoothed properly and all the ends of the branches were opened.
The local smoothing tool was applied with a proper smoothing strength, to further improve the finishing of the 3D models. The entire process of lung 3D model generation is depicted in Fig. 3, in the case of Patient 4.
At the end of these procedures, manual and automatic segmentation results were compared, in terms of time consumption and reconstruction accuracy and by extracting the number of identified airway branches. (This last comparison was limited to healthy subjects.) Moreover, the volume of both the left and the right lungs was acquired, together with the volume of the identified CT manifestations, to evaluate the global impact of the disease, for Patients 3–6. CT findings are classified according to Table 1.
Results
The required time for the overall manual segmentation process was approximately 9 h for each patient. The extraction of the airways was the most time demanding process (7 h), and adequate anatomy knowledge and experience were needed. The use of the automatic tools allowed a much faster identification of the airways (approximately 5 min), providing a good level of accuracy. The post-processing operations required nearly 1 h, in both cases. Regarding the manual segmentation of the lungs, about 1 h was needed, also including the post-processing operations. The automatic tools did not succeed in giving satisfactory results in COVID-19 positive patients and about 3 h of manual editing were necessary to obtain the final 3D reconstruction of the lungs. The most time-consuming operation, however, was the segmentation of the COVID-19 CT manifestations, with variable time consumption between 5 and 10 h.
In Fig. 4a, the final segmentation of the respiratory system of Patient 1 and Patient 2 is reported. A comparison of the manual identification of the airways and automatic algorithm result of Patient 1 is shown in Fig. 4b. The properties of 3D models are listed in Table 3.
In Table 4, the properties of 3D models of patients affected by COVID-19 are listed. In Fig. 5, the final segmentations of the respiratory system of Patients 3 to 6 are reported. Different colours were arbitrarily assigned to the abnormalities, to help visualization.
a 3D model of the respiratory system of Patient 3 (M, 70 years old, Typical appearance). b 3D model of the respiratory system of Patient 4 (F, 84 years old, Typical appearance). c 3D model of the respiratory system of Patient 5 (F, 33 years old, Indeterminate appearance). d 3D model of the respiratory system of Patient 6 (M, 60 years old, Indeterminate appearance)
In Table 5, the CT manifestations identified in each case are detailed, together with the Radiological Society of North America (RSNA) classification. The percentage of involvement of the lungs was evaluated, to assess the degree of diffusion of the disease.
Now, a sketched discussion about obtained reconstructions for COVID-19 positive patients is provided, illustrating Table 5 classification.
Patient 3 (male, 70 years old) (Fig. 5; Table 5): Patient 3 presented with predominantly peripheral bilateral GGO formations with rounded morphology, affecting different lung sections. The abnormalities found in the CT scan led to the RSNA classification as Typical appearance. Fibrous stripes, typical of the healing phase of the pulmonary chronic inflammation, were also observed close to the pleura. The percentage of the whole lung affected volume suggested a Moderate severity of the disease according to the literature [23] and, together with the presence of fibrosis, it may be indicative of a late stage of the viral infection [4, 24].
Patient 4 (female, 84 years old) (Fig. 5; Table 5): Patient 4 displayed signs of rounded GGOs, with the presence of crazy-paving pattern, reticular pattern, halo sign, consolidation, pleural thickening and fibrous stripes, leading to a Typical appearance classification. The high percentage of affected lung regions and the typology of CT findings may be representative of a peak stage of infection and of a Severe case.
Patient 5 (female, 33 years old) (Fig. 5; Table 5): Patient 5 showed non-rounded GGOs mainly in the left lung, lacking a specific distribution, crazy-paving pattern, consolidation and pleural thickening (Indeterminate appearance). Considering the extension of the affected lung region and the CT abnormalities, the infection was classified as Moderate type at the peak stage.
Patient 6 (male, 60 years old) (Fig. 5; Table 5): Patient 6 was labelled as Indeterminate appearance due to the non-rounded GGOs without a specific distribution and reticular pattern. This was classified as Severe disease in a progressive stage.
Discussion and conclusions
With both manual and automatic methods, it was possible to obtain a realistic reconstruction of the lower respiratory tract and of the pulmonary parenchyma. These 3D models could provide the clinicians with an overall view of the impact of the infection. Furthermore, a volumetric quantification of the CT findings can be performed, helping in the evaluation of the progression of the pathology and of the most proper therapeutic actions. In addition, 3D models provide a powerful tool for communicating the severity of COVID-19.
The use of an automatic algorithm allowed for a substantial reduction in the operating time in the case of airways segmentation; nonetheless, the number of identified branches was lower, in comparison with the detection by the operator through the visual inspection of the CT images. However, the model of the airways obtained with the automatic technique resulted to be more than adequate, if the assessment of COVID-19 impact on the respiratory system is considered the main target. In the case of healthy patients, the manual process of lung segmentation was not particularly time-consuming, nor complex. Whereas, for COVID-19 positive patients, the application of the automatic technique provided unacceptable results, requiring a laborious manual editing operation. Moreover, the greater the severity of the pathology, the worse the automatic segmentation result.
With respect to airway models, it was our observation that the accuracy of the detection was strongly affected by the CT imaging parameters. Comparing Patients 3 and 5, the slice thickness and the number of slices were the most relevant factors to be considered. In agreement with the literature, a low slice thickness determined an enhanced segmentation process [25]. Additionally, it was also observed that a higher number of slices led to a higher number of detected branches. Finally, with worsening levels of disease severity, poor segmentation was noted, despite similar CT scanning parameters, due to lower contrast between the airway walls and the pulmonary parenchyma.
To summarize, the most common COVID-19 CT findings in the patients analysed in this study were the GGOs and, in the case of severe pathology, the affected lung volume was mostly occupied by crazy-paving pattern and reticular pattern. It was also noticed that the infection typically involved the regions near the lung surfaces.
It is important to highlight certain limitations in this study. The present work was aimed at establishing a method to segment the pathological pulmonary manifestations in COVID-19 focusing just on four positive COVID-19 patients. To derive statistically significant data, it would be necessary to analyse a wider population ranging in age. Moreover, due to the lack of information on the clinical history of the analysed patients, it was not possible to avoid potential overlapping of the observed CT manifestations with those of other pathologies.
Anyway, we are confident the segmentation process that has been demonstrated could provide practising radiologists and researchers a tool to visualize, in a composite view, the many different pathological processes that take place in COVID-19 pneumonia, as well as their extent. Chest CTs in patients with active COVID-19 pneumonia are often obtained to assess complications and severity. Their association with 3D segmentation can provide clinicians with a comprehensive “snapshot” of the overall current status of pulmonary involvement that may prove to be useful in patient management. The categories of CT findings in COVID-19 pneumonia have been linked to temporal changes in pathology, from early to late radiological findings. Future studies, analysing the changes in the 3D segmentation images in individual patients over time, may help elucidate, in a more intuitive fashion, the nature of the progression of COVID-19 disease.
References
Hu B, Guo H, Zhou P, Shi ZL (2020) Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. https://doi.org/10.1038/s41579-020-00459-7
Sohrabi C, Alsafi Z, O’Neill N, Khan M, Kerwan A, Al-Jabir A, Iosifidis C, Agha R (2020) World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int J Surg 76:71–76. https://doi.org/10.1016/J.IJSU.2020.02.034
Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC (2020) Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA J Am Med Assoc 324:782–793. https://doi.org/10.1001/jama.2020.12839
Kwee TC, Kwee RM (2020) Chest CT in COVID-19: What the radiologist needs to know. Radiographics 40:1848–1865. https://doi.org/10.1148/rg.2020200159
Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, Cui J, Xu W, Yang Y, Fayad ZA, Jacobi A, Li K, Li S, Shan H (2020) CT imaging features of 2019 novel coronavirus (2019-NCoV). Radiology 295:202–207. https://doi.org/10.1148/radiol.2020200230
Ye Z, Zhang Y, Wang Y, Huang Z, Song B (2020) Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur Radiol 30:4381–4389. https://doi.org/10.1007/s00330-020-06801-0
Schachner ER, Spieler B (2020) Three-dimensional (3D) lung segmentation for diagnosis of COVID-19 and the communication of disease impact to the public. BMJ Case Rep 13:19–21. https://doi.org/10.1136/bcr-2020-236943
Tzeng YS, Hoffman E, Cook-Granroth J, Maurer R, Shah N, Mansour J, Tschirren J, Albert M (2007) Comparison of airway diameter measurements from an anthropomorphic airway tree phantom using hyperpolarized 3He MRI and high-resolution computed tomography. Magn Reson Med 58:636–642. https://doi.org/10.1002/mrm.21285
Rosell J, Cabras P (2013) A three-stage method for the 3D reconstruction of the tracheobronchial tree from CT scans. Comput Med Imaging Graph 37:430–437. https://doi.org/10.1016/j.compmedimag.2013.07.003
Kiraly AP, Higgins WE, McLennan G, Hoffman EA, Reinhardt JM (2002) Three-dimensional human airway segmentation methods for clinical virtual bronchoscopy. Acad Radiol 9:1153–1168. https://doi.org/10.1016/S1076-6332(03)80517-2
Salito C, Barazzetti L, Woods JC, Aliverti A (2011) 3D airway tree reconstruction in healthy subjects and emphysema. Lung 189:287–293. https://doi.org/10.1007/s00408-011-9305-4
Sonka M, Park W, Huffman EA (1996) Rule-based detection of intrathoracic airway trees. IEEE Trans Med Imaging 15:314–326. https://doi.org/10.1109/42.500140
Lo P, Van Ginneken B, Reinhardt JM, Yavarna T, De Jong PA, Irving B, Fetita C, Ortner M, Pinho R, Sijbers J, Feuerstein M, Fabijanska A, Bauer C, Beichel R, Mendoza CS, Wiemker R, Lee J, Reeves AP, Born S, Weinheimer O, Van Rikxoort EM, Tschirren J, Mori K, Odry B, Naidich DP, Hartmann I, Hoffman EA, Prokop M, Pedersen JH, De Bruijne M (2012) Extraction of airways from CT (EXACT’09). IEEE Trans Med Imaging 31:2093–2107. https://doi.org/10.1109/TMI.2012.2209674
Aykac D, Huffman EA, McLennan G, Reinhardt JM (2003) Segmentation and analysis of the human airway tree from three-dimensional X-ray CT images. IEEE Trans Med Imaging 22:940–950. https://doi.org/10.1109/TMI.2003.815905
Park W, Huffman EA, Sonka M (1998) Segmentation of intrathoracic airway trees: a fuzzy logic approach. IEEE Trans Med Imaging 17:489–497. https://doi.org/10.1109/42.730394
Ukil S, Reinhardt JM (2009) Anatomy-guided lung lobe segmentation in X-ray CT images. IEEE Trans Med Imaging 28:202–214. https://doi.org/10.1109/TMI.2008.929101
van Rikxoort E, Baggerman W, Ginneken B (2009) Automatic segmentation of the airway tree from thoracic CT scans using a multi-threshold approach. Second Work Pulm Image Anal: 341–349
Adams HJA, Kwee TC, Yakar D, Hope MD, Kwee RM (2020) Chest CT imaging signature of coronavirus disease 2019 infection. In pursuit of the scientific evidence. Chest 158:1885–1895. https://doi.org/10.1016/j.chest.2020.06.025
Simpson S, Kay FU, Abbara S, Bhalla S, Chung JH, Chung M, Henry TS, Kanne JP, Kligerman S, Ko JP, Litt H (2020) Radiological society of north America expert consensus statement on reporting chest CT findings related to COVID-19. endorsed by the society of thoracic radiology, the American college of radiology, and RSNA - secondary publication. J Thorac Imaging 35:219–227. https://doi.org/10.1097/RTI.0000000000000524
Rebouças Filho PP, Cortez PC, da Silva Barros AC, Victor VH, Tavares RSJM (2017) Novel and powerful 3D adaptive crisp active contour method applied in the segmentation of CT lung images. Med Image Anal 35:503–516. https://doi.org/10.1016/j.media.2016.09.002
Tsai EB, Simpson S, Lungren MP, Hershman M, Roshkovan L, Colak E, Erickson BJ, Shih G, Stein A, Kalpathy-Cramer J, Shen J, Hafez M, John S, Rajiah P, Pogatchnik BP, Mongan J, Altinmakas E, Ranschaert ER, Kitamura FC, Topff L, Moy L, Kanne JP, Wu CC (2021) The RSNA international COVID-19 open radiology database (RICORD). Radiology 299:E204–E213. https://doi.org/10.1148/radiol.2021203957
Noma S, Khan A, Herman PG, Rojas KA (1990) High-resolution computed tomography of the pulmonary parenchyma. Semin Ultrasound CT MR 11:365–379
Huang L, Han R, Ai T, Yu P, Kang H, Tao Q, Xia L (2020) Serial quantitative chest CT assessment of COVID-19: a deep learning approach. Radiol Cardiothorac Imaging 2:e200075. https://doi.org/10.1148/ryct.2020200075
Yu N, Shen C, Yu Y, Dang M, Cai S, Guo Y (2020) Lung involvement in patients with coronavirus disease-19 (COVID-19): a retrospective study based on quantitative CT findings. Chinese J Acad Radiol 3:102–107. https://doi.org/10.1007/s42058-020-00034-2
Nardelli P, Khan KA, Corvò A, Moore N, Murphy MJ, Twomey M, O’Connor OJ, Kennedy MP, Estépar RSJ, Maher MM, Cantillon-Murphy P (2015) Optimizing parameters of an open-source airway segmentation algorithm using different CT images. Biomed Eng Online 14:1–24. https://doi.org/10.1186/s12938-015-0060-2
Funding
Open access funding provided by Politecnico di Milano within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Human or Animal Rights
This article does not contain any studies with animals performed by any of the authors.
Informed consent
For this type of study, formal consent is not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bertolini, M., Brambilla, A., Dallasta, S. et al. High-quality chest CT segmentation to assess the impact of COVID-19 disease. Int J CARS 16, 1737–1747 (2021). https://doi.org/10.1007/s11548-021-02466-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02466-2