Abstract
Purpose
Intrauterine foetal surgery is the treatment option for several congenital malformations. For twin-to-twin transfusion syndrome (TTTS), interventions involve the use of laser fibre to ablate vessels in a shared placenta. The procedure presents a number of challenges for the surgeon, and computer-assisted technologies can potentially be a significant support. Vision-based sensing is the primary source of information from the intrauterine environment, and hence, vision approaches present an appealing approach for extracting higher level information from the surgical site.
Methods
In this paper, we propose a framework to detect one of the key steps during TTTS interventions—ablation. We adopt a deep learning approach, specifically the ResNet101 architecture, for classification of different surgical actions performed during laser ablation therapy.
Results
We perform a two-fold cross-validation using almost 50 k frames from five different TTTS ablation procedures. Our results show that deep learning methods are a promising approach for ablation detection.
Conclusion
To our knowledge, this is the first attempt at automating photocoagulation detection using video and our technique can be an important component of a larger assistive framework for enhanced foetal therapies. The current implementation does not include semantic segmentation or localisation of the ablation site, and this would be a natural extension in future work.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
Twin-to-twin transfusion syndrome (TTTS) is a disease affecting identical twin pregnancies. It is caused by abnormal vessels in the placenta that disproportionately transfuse blood from one twin to the other. The recipient of excessive blood is at risk of significant complications including heart failure, while the second foetus is affected by slower than normal growth. The overall outcome is dismal for both if the condition is left untreated. TTTS can be treated by coagulating the abnormal vessels in the placenta, interrupting the excessive blood flow from one twin to the other. Foetal surgery for TTTS involves the fetoscopic identification of anastomosing vessels on the unique twin placenta and laser photocoagulation [14]. However, the current procedure has several significant challenges: inability for correct orientation of endoscope and laser in case of anterior placenta [4, 9]; incomplete visualisation in case of turbid amniotic fluid or small vessels [11]; uncertainties with classification of arteries and veins based on oxygen content to guide sequential lasering [12]; instability of the image without motion compensation when carried out at close proximity to prevent inadvertent ablation or tissue contact and fetoplacental haemorrhage. Computer-assisted interventions (CAI) can offer potential solutions to some of these challenges. To achieve this, it is essential to use algorithms for extracting information from the the fetoscope, which is the only sensor within the in utero environment.
TTTS laser ablation therapy. a This procedure aims at coagulating a series of abnormal vessels in the placenta using a laser ablation tool inserted on a fetoscopic camera b fetoscopic image with the laser ablation tool visible and the placenta in the background; c timeline of a TTTS laser ablation procedure
TTTS interventions are performed using a fetoscopic camera with a retractable fibre-based laser ablation tool driven through a small working channel in the fetoscope. The surgeon navigates the fetoscope to sites on the placenta, identifying vascular structures and successively photocoagulating each of the abnormal vessels and targets them with the laser (Fig. 1c). The precise site for applying energy with the laser is controlled by the pose of the fetoscope as well as the insertion depth of the laser fibre. When inserted sufficiently, the ablation tool tip can be observed at the edge of the fetoscopic image as shown in Fig 1b. To assist the surgeon and indicate the point of energy delivery, the laser emits a low power projection that enables the surgeon to accurately aim at target vessels. Despite the fact that focused light from the laser ablation fibre provides very salient image features, especially in proximity with the placenta surface, there is a very high variability in image appearance between different patients, fetoscope hardware models, and different placental regions in intra- and inter-patients. This makes automatic video processing a challenge [2, 5, 6, 13] because complex appearance can occur due to a number of cases, for example: the fetoscope is not focused due to very dynamic changes in depth within the scene (Fig. 2a); the surgery is performed within a highly dynamic environment with foetuses that can move unpredictably and often occlude the camera field of view (Fig. 2b); the surgical environment is immersed in amniotic fluid, which becomes more turbid as the gestation evolves and degrades visibility (Fig. 2c); additionally, there is a very high variability in fetoscope light sources that can range from intense light with specularities on the placenta surface, to very dim lighting conditions (Fig. 2d). These factors render vision driven analysis of fetoscopic video a significant challenge and make it difficult to automatically detect events of interest, such as when the laser is actively photocoagulating.
Deep learning architectures for classification and semantic segmentation have been shown to be extremely effective in recent years. In minimally invasive surgery (MIS), such approaches have demonstrated to be the most effective for detection and classification of surgical instruments [7, 10] and even for detecting abnormal tissue structures [1] or segmenting surgical video into semantic steps [15]. In this paper, we take a similar approach and leverage the power of convolutional neural networks (CNNs) and their generalisation capabilities to handle high image variability in order to detect ablation during TTTS. This is the first time automation of recognition has been attempted in fetoscopic video.
Our contributions can be summarised as follows:
-
We design CNN-based methods for detecting the occurrence of ablation in fetoscopy videos while taking into account the specificities of TTTS ablation therapy and its impact on the characteristics of image appearance.
-
We demonstrate the generalisation ability of our method by testing it on large sequences with appearance characteristics that are significantly different to the training data.
-
We use our trained CNN to indirectly infer the occurrence of other events during the surgical procedure that would not allow ablation to occur in the immediate future, including occlusions, large distance to the placenta, and inactive state of laser tool. This information is obtained without the necessity of annotating these events in the training data and can be used to increase the accuracy of ablation detection.
Problem formulation
In this paper, our aim is to identify ablation during fetoscopic video and use this result to automatically segment different actions performed during a TTTS laser surgery using fetoscopic videos of the whole procedure. More specifically, we propose to separate surgical activity into three different classes. Targeting: The fetoscope is in close proximity to the placenta and could possibly start performing ablation in the immediate future. The laser ablation tool projects a green light circle on the surface of the placenta which indicates where its currently aiming at. Ablation: A vessel is being coagulated. The projected laser light increases in intensity and changes to a blue colour. The targeted vessel progressively changes in appearance, turning into a white coagulated blob. Other: Scenes where laser ablation could not possibly occur in the immediate future. The ablation tool is not targeting any surface, due to the fetoscope being too far from the placenta, occlusions, or the laser tool being turned off.
The most easily distinguishable label is ablation due to the blue light emission and the changes in appearance of the targeted vessel. During targeting, the laser emits a green light that is projected on the placenta. However, the distinction between targeting and other is not clear-cut (Fig. 3), as the surgeon may move the fetoscope away from the placenta without turning off the green laser pointer. In such cases, it becomes difficult to establish objective criteria to separate the two labels. This poses the additional problem of how to objectively annotate groundtruth data that belong to the targeting and other classes while avoiding ambiguous training labels.
There is temporal information that might help performing this task: the considered actions should span over consecutive sequences of frames to guarantee temporal continuity; ablation sequences should be temporally adjacent to targeting sequences; changes in appearance of the placenta surface might indicate the occurrence of ablation; and finally, the motion patterns of the laser tool and projected light are significantly different from the scene motion. However, in this paper we only consider classification based on the appearance of each frame independently.
Methods
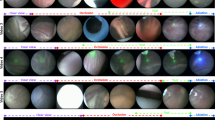
Our study considers videos obtained from five different TTTS ablation procedures (Fig. 4). The sequences have significantly different appearances between them: sequences 1 and 2 have relatively bright lighting, sequence 3 has a higher turbidity than normal, sequence 4 has a weaker light source, and in sequence 5 the projected light from the laser pointer is not visible in most of the frames (ablation is still noticeable from appearance changes in the ablation tool itself).
In order to avoid the labelling ambiguity mentioned in the previous section, we manually annotate the data with only two labels: ablation and \(\sim \)ablation. These are easily distinguishable by visually inspecting the colour and intensity of the laser emitted light and are relatively fast to perform on thousands of frames. We annotated a total of 49527 frames. A complete TTTS ablation procedure contains a significantly larger amount of \(\sim \)ablation frames than ablation frames. This class imbalance, if not properly addressed, causes a classification bias towards \(\sim \)ablation labels. We therefore randomly subsample the complete sequences in order to create training datasets with an equal number of ablation and \(\sim \)ablation labels. Figure 4 details the number of training frames extracted from each sequence.
When the laser ablation tool is exposed its orientation on the fetoscopic images will broadly remain the same for a single ablation procedure. However, this fixed orientation changes arbitrarily between different procedures. Our training datasets include sequences from few procedures, and since we do not want our algorithm to over-fit the label classification to these particular orientations of the ablation tool, we perform a random rotation on all images of our training datasets.
Binary classification network
We first consider the simpler problem of binary classification between ablation and \(\sim \)ablation labels.
We use the Residual Network (ResNet) architecture [8], which won the 2015 championship on three ImageNet competitions image classification, object localisation, and object detection. ImageNet [3] is a computer vision classification dataset containing non-medical images belonging to 1000 different labels. While previous CNN architectures usually suffer from over-fitting and slow convergence issues if the number of convolution layers is too high, ResNet has the ability to adapt well to the dimensionality and complexity of different problems while maintaining a high number of convolution layers. This makes it suitable to our problem, since we want to use a well performing CNN initialised on a large-scale dataset with very high variability, and adapt it to a problem with with only 2 labels and that deals with datasets that are limited in its appearance to laser ablation fetoscopy scenes.
We use the publicly available ResNet with 101 layers pre-trained on ImageNet as an initialisation (Fig. 5a). We then fine-tune the weights of all layers in this network using our training datasets. This network works with \(224\times 224\) input images; however, our captured fetoscopic images are significantly larger. We test different approaches to deal with the image size difference: downsampling the input images to a \(224\times 224\) resolution; or resizing them to a \(448\times 448\) resolution (very close to their original size) and modifying the final ResNet pooling layer so that it properly connects the higher resolution images to the final fully connected (FC) classification layer. We test two different pooling modifications: increasing the kernel size of the ResNet averaging pooling layer; maintaining the kernel size of ResNet’s averaging pooling layer and adding a max pooling layer. Note that this final modification acts as a single pooling layer that performs a maximum over averages operation.
For both approaches, the fully connected (FC) final layer of the original ResNet is replaced to output two labels instead of 1000. The network is fine-tuned for a duration of 30000 iterations, with a learning rate of \(1\times 10^{-5}\) and a batch size of four images.
Complete classification pipeline
In order to further separate the \(\sim \)ablation labels into targeting and other, we must first establish a definition for these labels. We propose to define targeting frames as being images with the same appearance as the ablation frames, except all the salient blue features are green instead. In other words, a targeting image is such that if we swap its blue and green channels, it would be classified as ablation, if given as input to an algorithm that is able to reliably estimate ablation labels. Therefore, our complete classification algorithm consists in running the same binary classification CNN on both the original image and its version with swapped green and blue channels, ending up with two binary classifications for each frame: {ablation,\(\sim \)ablation} and {targeting,\(\sim \)targeting}. If an image is classified as both \(\sim \)ablation and \(\sim \)targeting we attribute it the label other. If an image is classified as both ablation and targeting we attribute it the label with the maximum ResNet score. Note that this last step has an impact on the results of the binary classifier by re-labelling some ablation frames to \(\sim \)ablation. In the experimental section, we show that the re-labelled frames are mostly false positive ablation detections, and therefore, our complete classification pipeline has a filtering action that provides a higher accuracy in ablation detection than a binary classifier.
Results
We compare different ResNet-based algorithms for ablation detection and compare it against more classic SVM classification to demonstrate the non-trivial nature of this classification problem. The following algorithms are tested:
-
SVM+Hist Support vector machine (SVM) classifier with a Gaussian kernel function. We discretise the HSV colour space into 4096 bins (\(16\times 16 \times 16\)) and perform PCA to reduce image features to the most discriminant 1000 dimensions. We also tried to remove the V component before building histograms, to test whether ablation appearance is independent from the overall scene illumination intensity. However, this lead to worse detection results.
-
ResNet+AVG CNN binary classifier that increases the kernel size of the final average pooling layer of the ResNet architecture and uses \(448 \times 448\) images as input.
-
ResNet+MAX CNN binary classifier that adds an extra max pooling layer to the ResNet architecture and uses \(448 \times 448\) images as input.
-
DS+ResNet CNN binary classifier that downsamples input images to a \(224 \times 224\) resolution and uses the original ResNet architecture.
-
Filtered ResNet Our complete classification pipeline as described in Sect. 3.2. Note that this method can use any of the previous three algorithms as a basis for binary classification (Filtered ResNet+AVG, Filtered ResNet+MAX, Filtered DS+ResNet).
We use leave-one-out cross-validation to test the classification algorithms on each of the five datasets. The classification performance of the different methods is quantified in Table 1 by their precision (p), recall (r), and F-measure \(f_{1}= 2/( 1/r + 1/p)\). In this paper, we use the \(f_{1}\) an overall performance metric since it weights both precision and recall.
Binary classification
From Table 1, we can observe that SVM+Hist is only able to perform well on sequence 4. Between the basic binary CNN classifiers, ResNet+AVG provides the best results. The best performance is generally obtained on sequence 4 which is characterised by its dark lighting conditions. This makes the ablation laser light the dominant feature in the image. On the other hand, the worst case is sequence 5 in which the ablation laser light projection is too weak to be visibly projected on the placenta surface.
Complete classification
The filtering approach based on our complete classification pipeline improves all three CNN methods on dataset 2 and ResNet+MAX on dataset 4. On the remaining cases, the performance is either the same or marginally improved. Results are improved by increasing the precision at the cost of a slight decrease in recall, which is to be expected due to its filtering action on ablation labels. We also display in Fig. 6 the precision-recall curve for the three CNN-based algorithms, using the final labelling ResNet score as a varying parameter. It can be seen that filtering improves the overall performance of all CNN-based methods. Table 2 presents the cumulative results of the filtering approaches over all five validation sequences. Figure 7 displays a timeline with the filtered ResNet+MAX predictions of ablation and \(\sim \)ablation labels for the whole duration of all five sequences, representing both correct and incorrect detections. In Fig. 8, we display some examples of incorrect detections.
We note that this increase in performance is obtained while simultaneously identifying a third label that enhances the qualitative descriptiveness of TTTS ablation timelines. Figure 9 displays results for the classification of the three labels: ablation, targeting, and other. We notice that in the two best performing sequences, 3 and 4, most of the \(\sim \)ablation frames are labelled as targeting with a few short sequences of other labels being spread throughout the sequences. On the other hand, the worst performing sequence (5) presents the largest sequences of other labels.
Zoomed in details from sequence 4 in Fig. 9: beginning of the ablation procedure above, and end of the ablation procedure below
Given that sequence 4 is the one with the most accurate ablation labels, we examine it in more detail to analyse the distinctions between the labels targeting and other, and understand to what type of events they correspond to in the TTTS ablation procedure. Figure 10 displays in more detail the beginning and the end of sequence 4. At the beginning of the procedure one can notice that before the laser is turned on (image a) the frames are classified as other and as soon as the laser is turned on the frames are classified as targeting (image b). It is also worthy of notice that a small sequence of other labels (image e) is due to the scene being occluded by the umbilical chord. At the end of the procedure, we can further analyse the behaviour of our classification network. There is a series of intermittent classifications between targeting and targeting which indicates an ambiguous sequence of frames. This is caused by the fetoscope being further away from the placenta (Image 1). The transition between Image 2 and Image 3 is caused by an occlusion with the umbilical chord. In image 4, the fetoscope is again starting to move away from the placenta causing the laser projection to be progressively less focused. When the fetoscope is furthest away from the placenta (Image 5), the laser pattern is significantly less visible. Image 6 is an easy classification example where the laser is clearly visible again. Image 7 displays a case where the fetoscope is very close to the placenta, but the laser light is not visible due to the angle at which the fetoscope is positioned. Image 8 belongs to the very ending of the procedure where the camera slowly moves away from the placenta until it is completely removed from the patient. The laser only focuses on the placenta for very short periods of time (Image 9), but for the most part the laser is not visible at all (Image 10). Image 11 depicts an interesting sequence where the camera is removed from the patient through an entirely blue cannula, causing a few false positive detections.
Discussion
From our results we can observe that a simple SVM solution based on colour histograms is not enough to predict ablation labels. All CNN-based methods perform better with darker environments (sequence 4), and worst with weak ablation laser lighting (sequence 5). Additionally, our complete pipeline seems to detect more targeting labels in datasets where the ablation laser is more visible. On one hand, this makes sense since we expect that the tool is mostly ready for ablation once its laser light is clearly projected on the placenta, showing sufficient proximity. On the other hand, this might not account for cases where the surgeon has to deal with poor visibility conditions throughout the whole procedure. This is an important information to take into account when selecting data for future expansions of the training dataset.
The fact that the Filtered ResNet approaches provide the best results shows that our heuristic approach to distinguish between targeting and other labels produces meaningful results. Our established prior that images similar to ablation with a green pattern belong to a label on its own provides useful information that increases the accuracy of ablation detection without the need of additional image annotation. This is further confirmed by the qualitative analysis on Fig. 10 where we show that, when ablation detection is accurate, other labels correspond to meaningful events (occlusions, laser turned off, camera too distant from placenta, etc). Obviously, we expect that in datasets where ablation detection is less successful (sequence 5), the frames classified as other will be also less meaningful and descriptive.
Conclusions
In this paper, we have shown that automatic detection of surgical action in fetoscopic video is feasible. Our focus was on ablation detection from video-based analysis as a precursor capability for more advanced CAI capabilities. While hardware solutions may be possible, a pure vision approach is appealing within the context of broader visual scene understanding. This problem is seemingly easy to solve due to the emission of a coloured light from the laser fibre but is not the case in practice due to the complexity of the environment during TTTS. Therefore, simple approaches such as classification on colour histograms failed to provide any meaningful results.
Our results indicate that the deep learning approaches perform beyond the capabilities of hand crafted techniques for an environment that presents many dynamic challenges. We also show that it is possible to incorporate heuristic prior information to increase detection accuracy and potentially provide useful information to distinguish between finer segmentation labels. Additionally, the recognition of different vessel regions, the detection of coagulated vessels, as well as motion analysis could potentially be useful to distinguish between actual targeting periods immediately before an ablation and other different tasks that are very similar in image appearance, such as vessel inspection after ablation, and navigation between two target vessels. We also need to experiment further with different types of scopes and ablation tools in order to further expand the training set.
References
Bernal J, Tajkbaksh N, Snchez FJ, Matuszewski BJ, Chen H, Yu L, Angermann Q, Romain O, Rustad B, Balasingham I, Pogorelov K, Choi S, Debard Q, Maier-Hein L, Speidel S, Stoyanov D, Brandao P, Crdova H, Snchez-Montes C, Gurudu SR, Fernndez-Esparrach G, Dray X, Liang J, Histace A (2017) Comparative validation of polyp detection methods in video colonoscopy: results from the miccai 2015 endoscopic vision challenge. IEEE Trans Med Imaging 36(6):1231–1249
Daga P, Chadebecq F, Shakir DI, Herrera LCGP, Tella M, Dwyer G, David AL, Deprest J, Stoyanov D, Vercauteren T, Ourselin S (2016) Real-time mosaicing of fetoscopic videos using sift. In: Proceedings of SPIE, SPIE medical imaging, vol 9786
Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L (2009) Imagenet: a large-scale hierarchical image database. In: IEEE conference on computer vision and pattern recognition, CVPR 2009, pp 248–255
Deprest J, Van Schoubroeck D, Van Ballaer P, Flageole H, Van Assche FA, Vandenberghe K (1998) Alternative technique for Nd: YAG laser coagulation in twin-to-twin transfusion syndrome with anterior placenta. Ultrasound in Obstet Gynecol 11(5):347–352
Gaisser F, Jonker PP, Chiba T (2016) Image registration for placenta reconstruction. In: Proceedings of the IEEE conference on computer vision and pattern recognition workshops, pp 33–40
Gaisser F, Peeters SHP, Lenseigne B, Jonker PP, Oepkes D (2017) Fetoscopic panorama reconstruction: moving from ex-vivo to in-vivo. Springer, Cham, pp 581–593. https://doi.org/10.1007/978-3-319-60964-5_51
Garcia-Peraza-Herrera LC, Li W, Fidon L, Gruijthuijsen C, Devreker A, Attilakos G, Deprest J, Vander Poorten E, Stoyanov D, Vercauteren T, Ourselin S (2017) Toolnet: holistically-nested real-time segmentation of robotic surgical tools. In: Proceedings of the 2017 IEEE/RSJ international conference on intelligent robots and systems
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Huber A, Baschat A, Bregenzer T, Diemert A, Tchirikov M, Hackelöer B, Hecher K (2008) Laser coagulation of placental anastomoses with a 30 fetoscope in severe mid-trimester twin-twin transfusion syndrome with anterior placenta. Ultrasound Obstet Gynecol 31(4):412–416
Kurmann T, Neila PM, Du X, Fua P, Stoyanov D, Wolf S, Sznitman R (2017) Simultaneous recognition and pose estimation of instruments in minimally invasive surgery. In: International conference on medical image computing and computer-assisted intervention, Springer, Berlin. pp 505–513
Lewi L, Deprest J, Hecher K (2013) The vascular anastomoses in monochorionic twin pregnancies and their clinical consequences. Am J Obstet Gynecol 208(1):19–30. https://doi.org/10.1016/j.ajog.2012.09.025
Quintero RA, Ishii K, Chmait RH, Bornick PW, Allen MH, Kontopoulos EV (2007) Sequential selective laser photocoagulation of communicating vessels in twin-twin transfusion syndrome. J Maternal-Fetal Neonatal Med 20(10):763–768
Reeff M, Gerhard F, Cattin P, , Szkely G (2006) Mosaicing of endoscopic placenta images. In: Hochberger C, Liskowsky R (eds) Informatik 2006. Informatik fr Menschen, Lecture Notes in Infomatics, vol P-93, pp 467–474
Senat MV, Deprest J, Boulvain M, Paupe A, Winer N, Ville Y (2004) Endoscopic laser surgery versus serial amnioreduction for severe twin-to-twin transfusion syndrome. New Engl J Med 351(2):136–144
Twinanda AP, Shehata S, Mutter D, Marescaux J, de Mathelin M, Padoy N (2017) Endonet: a deep architecture for recognition tasks on laparoscopic videos. IEEE Trans Med Imaging 36(1):86–97
Acknowledgements
This work was supported through an Innovative Engineering for Health award by the Wellcome Trust [WT101957] and an Engineering and Physical Sciences Research Council (EPSRC) [NS/A000027/1].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
For this type of study formal consent is not required.
Informed consent
No animals or humans were involved in this research. All videos were anonymised before delivery to the researchers.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Vasconcelos, F., Brandão, P., Vercauteren, T. et al. Towards computer-assisted TTTS: Laser ablation detection for workflow segmentation from fetoscopic video. Int J CARS 13, 1661–1670 (2018). https://doi.org/10.1007/s11548-018-1813-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-018-1813-8