Abstract
Purpose
To assess the efficacy of machine learning and radiomics analysis by computed tomography (CT) in presurgical setting, to predict RAS mutational status in colorectal liver metastases.
Methods
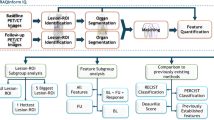
Patient selection in a retrospective study was carried out from January 2018 to May 2021 considering the following inclusion criteria: patients subjected to surgical resection for liver metastases; proven pathological liver metastases; patients subjected to enhanced CT examination in the presurgical setting with a good quality of images; and RAS assessment as standard reference. A total of 851 radiomics features were extracted using the PyRadiomics Python package from the Slicer 3D image computing platform after slice-by-slice segmentation on CT portal phase by two expert radiologists of each individual liver metastasis performed first independently by the individual reader and then in consensus. Balancing technique was performed, and inter- and intraclass correlation coefficients were calculated to assess the between-observer and within-observer reproducibility of features. Receiver operating characteristics (ROC) analysis with the calculation of area under the ROC curve (AUC), sensitivity (SENS), specificity (SPEC), positive predictive value (PPV), negative predictive value (NPV) and accuracy (ACC) were assessed for each parameter. Linear and non-logistic regression model (LRM and NLRM) and different machine learning-based classifiers were considered. Moreover, features selection was performed before and after a normalized procedure using two different methods (3-sigma and z-score).
Results
Seventy-seven liver metastases in 28 patients with a mean age of 60 years (range 40–80 years) were analyzed. The best predictors, at univariate analysis for both normalized procedures, were original_shape_Maximum2DDiameter and wavelet_HLL_glcm_InverseVariance that reached an accuracy of 80%, an AUC ≥ 0.75, a sensitivity ≥ 80% and a specificity ≥ 70% (p value < < 0.01). However, a multivariate analysis significantly increased the accuracy in RAS prediction when a linear regression model (LRM) was used. The best performance was obtained using a LRM combining linearly 12 robust features after a z-score normalization procedure: AUC of 0.953, accuracy 98%, sensitivity 96%, specificity of 100%, PPV 100% and NPV 96% (p value < < 0.01). No statistically significant increase was obtained considering the tested machine learning both without normalization and with normalization methods.
Conclusions
Normalized approach in CT radiomics analysis allows to predict RAS mutational status in colorectal liver metastases patients.




Similar content being viewed by others
Data availability
Data are reported in the manuscript and at link https://zenodo.org/records/10820402
References
Ottaiano A, Scala S, Santorsola M, Trotta AM, D’Alterio C, Portella L, Clemente O, Nappi A, Zanaletti N, De Stefano A, Avallone A, Granata V, Notariello C, Luce A, Lombardi A, Picone C, Petrillo A, Perri F, Tatangelo F, Di Mauro A, Albino V, Izzo F, Rega D, Pace U, Di Marzo M, Chiodini P, De Feo G, Del Prete P, Botti G, Delrio P, Caraglia M, Nasti G (2021) Aflibercept or bevacizumab in combination with FOLFIRI as second-line treatment of mRAS metastatic colorectal cancer patients: the ARBITRATION study protocol. Ther Adv Med Oncol 24(13):1758835921989223. https://doi.org/10.1177/1758835921989223
Ottaiano A, Caraglia M, Di Mauro A, Botti G, Lombardi A, Galon J, Luce A, D’Amore L, Perri F, Santorsola M, Hermitte F, Savarese G, Tatangelo F, Granata V, Izzo F, Belli A, Scala S, Delrio P, Circelli L, Nasti G (2020) Evolution of mutational landscape and tumor immune-microenvironment in liver oligo-metastatic colorectal cancer. Cancers (Basel) 12(10):3073. https://doi.org/10.3390/cancers12103073
Cervantes A, Adam R, Roselló S, Arnold D, Normanno N, Taïeb J, Seligmann J, De Baere T, Osterlund P, Yoshino T, Martinelli E, ESMO Guidelines Committee (2023) Electronic address: clinicalguidelines@esmo.org. Metastatic colorectal cancer: ESMO clinical practice guideline for diagnosis, treatment and follow-up. Ann Oncol 34(1):10–32. https://doi.org/10.1016/j.annonc.2022.10.003
Mateo J, Steuten L, Aftimos P, André F, Davies M, Garralda E, Geissler J, Husereau D, Martinez-Lopez I, Normanno N, Reis-Filho JS, Stefani S, Thomas DM, Westphalen CB, Voest E (2022) Delivering precision oncology to patients with cancer. Nat Med 28(4):658–665. https://doi.org/10.1038/s41591-022-01717-2
Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, Normanno N, Scarpa A, Robson M, Meric-Bernstam F, Wagle N, Stenzinger A, Bonastre J, Bayle A, Michiels S, Bièche I, Rouleau E, Jezdic S, Douillard JY, Reis-Filho JS, Dienstmann R, André F (2020) Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO precision medicine working group. Ann Oncol 31(11):1491–1505. https://doi.org/10.1016/j.annonc.2020.07.014
Martinelli E, Ciardiello D, Martini G, Troiani T, Cardone C, Vitiello PP, Normanno N, Rachiglio AM, Maiello E, Latiano T, De Vita F, Ciardiello F (2020) Implementing anti-epidermal growth factor receptor (EGFR) therapy in metastatic colorectal cancer: challenges and future perspectives. Ann Oncol 31(1):30–40. https://doi.org/10.1016/j.annonc.2019.10.007
Ottaiano A, Sabbatino F, Perri F, Cascella M, Sirica R, Patrone R, Capuozzo M, Savarese G, Ianniello M, Petrillo N, Circelli L, Granata V, Berretta M, Santorsola M, Nasti G (2023) KRAS p.G12C mutation in metastatic colorectal cancer: prognostic implications and advancements in targeted therapies. Cancers (Basel) 15(14):3579. https://doi.org/10.3390/cancers15143579
Collienne M, Neven A, Caballero C, Kataoka K, Carrion-Alvarez L, Nilsson H, Désolneux G, Rivoire M, Ruers T, Gruenberger T, Protic M, Troisi RI, Primavesi F, Staettner S, Rahbari N, Schnitzbauer A, Malik H, Swijnenburg RJ, Mauer M, Ducreux M, Evrard S (2023) EORTC 1409 GITCG/ESSO 01–A prospective colorectal liver metastasis database for borderline or initially unresectable diseases (CLIMB): lessons learnt from real life. From paradigm to unmet need. Eur J Surg Oncol. 49(11):107081. https://doi.org/10.1016/j.ejso.2023.107081
Vauthey JN, Zimmitti G, Kopetz SE, Shindoh J, Chen SS, Andreou A, et al. (2013) Ras mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg 258(4):619–26. discussion 26–7
Margonis GA, Sasaki K, Gholami S, Kim Y, Andreatos N, Rezaee N et al (2018) Genetic and morphological evaluation (game) score for patients with colorectal liver metastases. Br J Surg 105(9):1210–1220
Brudvik KW, Jones RP, Giuliante F, Shindoh J, Passot G, Chung MH et al (2019) Ras mutation clinical risk score to predict survival after resection of colorectal liver metastases. Ann Surg 269(1):120–126
Lambin P, Rios-Velazquez E, Leijenaar R, Carv-alho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446
Dercle L, Lu L, Schwartz LH, Qian M, Tejpar S, Eggleton P, Zhao B, Piessevaux H (2020) Radiomics response signature for identification of metastatic colorectal cancer sensitive to therapies targeting EGFR pathway. J Natl Cancer Inst 112(9):902–912. https://doi.org/10.1093/jnci/djaa017
Bera K, Braman N, Gupta A, Velcheti V, Madabhushi A (2022) Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat Rev Clin Oncol 19(2):132–146. https://doi.org/10.1038/s41571-021-00560-7
Taghavi M, Trebeschi S, Simões R, Meek DB, Beckers RCJ, Lambregts DMJ, Verhoef C, Houwers JB, van der Heide UA, Beets-Tan RGH, Maas M (2021) Machine learning-based analysis of CT radiomics model for prediction of colorectal metachronous liver metastases. Abdom Radiol (NY) 46(1):249–256. https://doi.org/10.1007/s00261-020-02624-1
Granata V, Fusco R, Barretta ML, Picone C, Avallone A, Belli A, Patrone R, Ferrante M, Cozzi D, Grassi R, Grassi R, Izzo F, Petrillo A (2021) Radiomics in hepatic metastasis by colorectal cancer. Infect Agent Cancer 16(1):39. https://doi.org/10.1186/s13027-021-00379-y
Petrillo A, Fusco R, Barretta ML, Granata V, Mattace Raso M, Porto A, Sorgente E, Fanizzi A, Massafra R, Lafranceschina M, La Forgia D, Trombadori CML, Belli P, Trecate G, Tenconi C, De Santis MC, Greco L, Ferranti FR, De Soccio V, Vidiri A, Botta F, Dominelli V, Cassano E, Boldrini L (2023) Radiomics and artificial intelligence analysis by T2-weighted imaging and dynamic contrast-enhanced magnetic resonance imaging to predict breast cancer histological outcome. Radiol Med. https://doi.org/10.1007/s11547-023-01718-2
Granata V, Fusco R, De Muzio F, Brunese MC, Setola SV, Ottaiano A, Cardone C, Avallone A, Patrone R, Pradella S, Miele V, Tatangelo F, Cutolo C, Maggialetti N, Caruso D, Izzo F, Petrillo A (2023) Radiomics and machine learning analysis by computed tomography and magnetic resonance imaging in colorectal liver metastases prognostic assessment. Radiol Med. https://doi.org/10.1007/s11547-023-01710-w
Granata V, Fusco R, De Muzio F, Cutolo C, Setola SV, Dell’Aversana F, Grassi F, Belli A, Silvestro L, Ottaiano A, Nasti G, Avallone A, Flammia F, Miele V, Tatangelo F, Izzo F, Petrillo A (2022) Radiomics and machine learning analysis based on magnetic resonance imaging in the assessment of liver mucinous colorectal metastases. Radiol Med 127(7):763–772. https://doi.org/10.1007/s11547-022-01501-9
Granata V, Fusco R, De Muzio F, Cutolo C, Mattace Raso M, Gabelloni M, Avallone A, Ottaiano A, Tatangelo F, Brunese MC, Miele V, Izzo F, Petrillo A (2022) Radiomics and machine learning analysis based on magnetic resonance imaging in the assessment of colorectal liver metastases growth pattern. Diagn (Basel) 12(5):1115. https://doi.org/10.3390/diagnostics12051115
Granata V, Fusco R, Setola SV, De Muzio F, Dell’ Aversana F, Cutolo C, Faggioni L, Miele V, Izzo F, Petrillo A (2022) CT-based radiomics analysis to predict histopathological outcomes following liver resection in colorectal Liver metastases. Cancers (Basel) 14(7):1648. https://doi.org/10.3390/cancers14071648
Granata V, Fusco R, De Muzio F, Cutolo C, Setola SV, Grassi R, Grassi F, Ottaiano A, Nasti G, Tatangelo F, Pilone V, Miele V, Brunese MC, Izzo F, Petrillo A (2022) Radiomics textural features by MR imaging to assess clinical outcomes following liver resection in colorectal liver metastases. Radiol Med 127(5):461–470. https://doi.org/10.1007/s11547-022-01477-6
Granata V, Fusco R, De Muzio F, Cutolo C, Setola SV, Dell’Aversana F, Ottaiano A, Nasti G, Grassi R, Pilone V, Miele V, Brunese MC, Tatangelo F, Izzo F, Petrillo A (2022) EOB-MR based radiomics analysis to assess clinical outcomes following liver resection in colorectal liver metastases. Cancers (Basel) 14(5):1239. https://doi.org/10.3390/cancers14051239
Granata V, Fusco R, De Muzio F, Cutolo C, Setola SV, Dell’ Aversana F, Ottaiano A, Avallone A, Nasti G, Grassi F, Pilone V, Miele V, Brunese L, Izzo F, Petrillo A (2022) Contrast MR-based radiomics and machine learning analysis to assess clinical outcomes following liver resection in colorectal liver metastases: a preliminary study. Cancers (Basel) 14(5):1110. https://doi.org/10.3390/cancers14051110
van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillon-Robin JC, Pieper S, Aerts HJWL (2017) Computational radiomics system to decode the radiographic phenotype. Can Res 77(21):e104–e107. https://doi.org/10.1158/0008-5472.CAN-17-0339
Kocak B, Akinci D’Antonoli T, Mercaldo N et al (2024) METhodological Radiomics Score (METRICS): a quality scoring tool for radiomics research endorsed by EuSoMII. Insights Imaging 15:8. https://doi.org/10.1186/s13244-023-01572-w
Kocak B, Baessler B, Bakas S, Cuocolo R, Fedorov A, Maier-Hein L, Mercaldo N, Müller H, Orlhac F, Pinto Dos Santos D, Stanzione A, Ugga L, Zwanenburg A (2023) CheckList for evaluation of radiomics research (CLEAR): a step-by-step reporting guideline for authors and reviewers endorsed by ESR and EuSoMII. Insights Imaging 14(1):75. https://doi.org/10.1186/s13244-023-01415-8
Chen Z, Lin T, Xia X, Xu H, Ding S (2017) A synthetic neighborhood generation based ensemble learning for the imbalanced data classification. Appl Intell 48:2441–2457
Tibshirani R (1996) Regression shrinkage and selection Via the Lasso. J R Stat Soc Ser B Statist Methodol. 58:267–288
Granata V, Fusco R, Avallone A, De Stefano A, Ottaiano A, Sbordone C, Brunese L, Izzo F, Petrillo A (2021) Radiomics-derived data by contrast enhanced magnetic resonance in RAS mutations detection in colorectal liver metastases. Cancers (Basel) 13(3):453. https://doi.org/10.3390/cancers13030453
Granata V, Fusco R, Risi C, Ottaiano A, Avallone A, De Stefano A, Grimm R, Grassi R, Brunese L, Izzo F, Petrillo A (2020) Diffusion-weighted MRI and diffusion kurtosis imaging to detect RAS mutation in colorectal liver metastasis. Cancers (Basel) 12(9):2420. https://doi.org/10.3390/cancers12092420
Scalco E, Belfatto A, Mastropietro A, Rancati T, Avuzzi B, Messina A, Valdagni R, Rizzo G (2020) T2w-MRI signal normalization affects radiomics features reproducibility. Med Phys 47(4):1680–1691. https://doi.org/10.1002/mp.14038
Granzier RWY, Ibrahim A, Primakov S, Keek SA, Halilaj I, Zwanenburg A, Engelen SME, Lobbes MBI, Lambin P, Woodruff HC, Smidt ML (2022) Test-retest data for the assessment of breast mri radiomic feature repeatability. J Magn Reson Imaging 56(2):592–604. https://doi.org/10.1002/jmri.28027
Jia LL, Zhao JX, Zhao LP, Tian JH, Huang G (2023) Current status and quality of radiomic studies for predicting KRAS mutations in colorectal cancer patients: a systematic review and meta-analysis. Eur J Radiol 158:110640. https://doi.org/10.1016/j.ejrad.2022.110640
Lubner MG, Stabo N, Lubner SJ, del Rio AM, Song C, Halberg RB, Pickhardt PJ (2015) CT textural analysis of hepatic metastatic colorectal cancer: pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom Imaging 40(7):2331–2337. https://doi.org/10.1007/s00261-015-0438-4
Shi R, Chen W, Yang B, Qu J, Cheng Y, Zhu Z, Gao Y, Wang Q, Liu Y, Li Z, Qu X (2020) Prediction of KRAS, NRAS and BRAF status in colorectal cancer patients with liver metastasis using a deep artificial neural network based on radiomics and semantic features. Am J Cancer Res 10(12):4513–4526
Mayerhoefer ME, Szomolanyi P, Jirak D, Materka A, Trattnig S (2009) Effects of MRI acquisition parameter variations and protocol heterogeneity on the results of texture analysis and pattern discrimination: an application-oriented study. Med Phys 36(4):1236–1243. https://doi.org/10.1118/1.3081408
Siriwardena AK, Serrablo A, Fretland ÅA, Wigmore SJ, Ramia-Angel JM, Malik HZ, Stättner S, Søreide K, Zmora O, Meijerink M, Kartalis N, Lesurtel M, Verhoef K, Balakrishnan A, Gruenberger T, Jonas E, Devar J, Jamdar S, Jones R, Hilal MA, Andersson B, Boudjema K, Mullamitha S, Stassen L, Dasari BVM, Frampton AE, Aldrighetti L, Pellino G, Buchwald P, Gürses B, Wasserberg N, Gruenberger B, Spiers HVM, Jarnagin W, Vauthey JN, Kokudo N, Tejpar S, Valdivieso A, Adam R (2023) Multisocietal European consensus on the terminology, diagnosis, and management of patients with synchronous colorectal cancer and liver metastases: an E-AHPBA consensus in partnership with ESSO, ESCP, ESGAR, and CIRSE. Br J Surg 110(9):1161–1170. https://doi.org/10.1093/bjs/znad124
Acknowledgements
The authors are grateful to Alessandra Trocino, librarian at the National Cancer Institute of Naples, Italy.
Funding
This work was supported by the Italian Ministry of Health Ricerca Corrente funds.
Author information
Authors and Affiliations
Contributions
Each author contributed to methodology, enrollment of patient and investigation. Each author written, revised and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Roberta Fusco and Vincenza Granata are editors in Radiologia Medica journal.
Ethical approval
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee National Cancer Institute of Naples Pascale Foundation.
Consent to participate
All patients enrolled signed the informed consent.
Consent to publish
Each author gives consent for publication of the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Granata, V., Fusco, R., Setola, S.V. et al. Machine learning and radiomics analysis by computed tomography in colorectal liver metastases patients for RAS mutational status prediction. Radiol med (2024). https://doi.org/10.1007/s11547-024-01828-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11547-024-01828-5