Abstract
High levels of resistance against Meloidogyne hapla have been identified in wild species of tuber-bearing potatoes, but only QTL with partial effects have been identified so far in back crosses with cultivated potato. This study was designed to test if pyramiding of two previously identified resistance genes, R Mh-tar and R Mh-chc A, will result in improved or even an absolute level of resistance. R Mh-tar and R Mh-chc A introgressed from the wild tuber-bearing potato species Solanum tarijense and Solanum chacoense were combined in a segregating diploid Solanum tuberosum population. With the aid of AFLP markers, descendants from this segregating population were classified into four groups, carrying no R gene, with only R Mh-tar , with only R Mh-chc A and a group with the pyramided R Mh-tar and R Mh-chc A. Upon inoculation with M. hapla isolate Bovensmilde, the group containing only R Mh-chc A showed a decline of 88% in average number of developed egg masses compared to the group without R Mh-chc A and R Mh-tar . The group of genotypes containing only R Mh-tar , but not R Mh-chc A, showed a decline of 55% in the number of developed egg masses compared to the group without R Mh-chc A and R Mh-tar . Unfortunately, the latter effect of R Mh-tar was not significant. The effect of both loci, R Mh-tar and R Mh-chc A combined, did not further reduce the number of egg masses compared to the level of R Mh-chc A alone.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Compared to other potato diseases and pests such as late blight or potato cyst nematodes, infection with the root knot nematode species Meloidogyne hapla is less noticeable. The damage caused by this pest in potato can be expressed only in yield reduction (Mugniéry 2007). However, in extremely infected soils, the yield reduction can amount up to 70% (Macguidwin and Rouse 1990). Crop rotation, as a strategy to reduce disease and pest pressure, is less practicable to control M. hapla infection because of its broad host spectrum. Meloidogyne hapla propagates on many dicotylenous plants and will therefore increasingly infect following crops (Mugniéry 2007).
While a number of resistance genes have been incorporated in cultivars, primarily against late blight, viruses, wart disease and potato cyst nematodes, no M. hapla-resistant cultivars are currently available. The deployment of cultivars resistant against cyst nematodes allowed suspending soil fumigation with nematicides. Therefore, a shift in the nematode populations towards free living and root knot nematodes such as M. hapla should be expected in the immediate future.
Introgression of resistance genes from wild Solanum species is viewed as an efficient solution to reduce M. hapla infection. A broad range of wild tuber-bearing Solanum species has been tested for M. hapla resistance by Janssen et al. (1996b). High levels of resistance have been identified in 14 different Solanum species. In these studies, it already became apparent that the resistance to M. hapla in these wild species would not lead to absolute immunity in potato. Draaistra (2006) studied the inheritance of M. hapla resistance originating from Solanum chacoense (accession BGRC 18618) and Solanum tarijense (accession BGRC 24717) and localized genetic loci involved in resistance on linkage maps. The major S. chacoense quantitative trait locus (QTL), named R Mh-chc A, explained 38% of the phenotypic variance, and mapped to a distal region of a linkage group that has not been assigned to a potato chromosome. From the same resistant parent, another QTL named R Mh-chc B descends. This small-effect QTL explained only 9.6% of the phenotypic variance and its contribution is ignored in this pyramiding study. The locus involved in resistance originating from S. tarijense (R Mh-tar ) explained 20% of the phenotypic variance and could be mapped as a qualitative trait locus to a distal location on potato chromosome 7. Offspring from neither S. chacoense nor S. tarijense displayed absolute levels of resistance, which is essential to avoid propagation of the pathogen population. At this moment, it is not fully understood why the transmission of resistance from highly resistant wild species only results in back cross offspring with partial resistance to M. hapla. This study was designed to test if pyramiding of these two resistance genes R Mh-chc A and R Mh-tar will result in improved or even an absolute level of resistance.
Materials and Methods
Plant Material
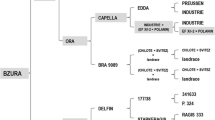
The diploid full-sib population RH04-456, pyramiding two loci involved in M. hapla resistance, was generated from a cross between clones RH90-011-4 and 87-206-6. Both parents were previously used as parents for the respective mapping populations that allowed the identification and genetic mapping of R Mh-tar and R Mh-chc A (Draaistra 2006). In the initial study, another small-effect QTL was identified, named R Mh-chc B, originating from S. chacoense parent 87-206-6 (Draaistra 2006), which is ignored in this study. The R Mh-tar donor RH90-011-4 was obtained from an interspecific cross between S. tarijense (BGRC 24717/CGN 18107) and MON-46, a dihaploid clone of the susceptible cultivar Mondial (see Fig. 1). The S. chacoense resistance from 87-206-6 was first described by Janssen et al. (1996a) and later by Draaistra (2006). Plant material was propagated to obtain sufficient seed tubers for the disease test.
Meloidogyne hapla Resistance Assay
One hundred twelve offspring genotypes of pyramiding population RH04-456 were tested for M. hapla resistance. Tubers were planted in 500-ml clay pots containing silversand and a slow release NPK fertiliser (Osmocote). The genotypes were tested for their level of resistance in three replications in a randomized complete block design. The susceptible cultivar Nicola and the resistant parents RH90-011-4 and 87-206-6 were included as control plants. Three weeks later, when the root systems were fully developed and reached through the bottom hole of the pots, the plants were inoculated with M. hapla isolate “Bovensmilde” at a concentration of 750 second stage juveniles (J2) per pot. The inoculum “Bovensmilde”, is commercially available from RZ Research, Metslawier, The Netherlands. The first 2 weeks after inoculation, the plants were watered with great care to avoid flushing the juveniles from the pots. Eight weeks after inoculation, the root systems were released from the sand by rinsing with tap water. Egg masses were stained with Phloxine-B (Dickson and Struble 1965) and counted. Variation in the size of the root system was recorded on a scale of 1–5, where 1 = small root system and 5 = large root system.
DNA Extraction
Genomic DNA from the parents and the offspring population RH04-456 was isolated from young leaf material essentially as described by Stewart and Via (1993). Leaf material was ground with steel balls using a Retsch machine (Retsch, Haan, Germany) in the extraction buffer, with volumes adapted to 2-ml-deep 96-well Costar plates (Corning Inc., Corning, NY, USA). DNA was visualised on a 1% agarose gel to check its integrity. DNA concentrations were measured with a NanoDrop™ ND 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Genotyping with AFLP™ Markers
The genetic maps constructed in the mapping studies of R Mh-chc A and R Mh-tar were solely constructed with AFLP™ markers (Draaistra 2006). Therefore, the same AFLP markers linked to R Mh-chc A and R Mh-tar were used to analyse the genetic composition of the offspring genotypes of the pyramiding population. AFLP was performed according to Vos et al. (1995). Fluorescently labelled AFLP fragments were visualised on a denaturing polyacrylamide gel using a NEN® Global Edition IR2 DNA Analyzer (LI-COR® Biosciences, Lincoln, NE, USA). The AFLP fragments were scored visually for absence/presence. Three primer combinations per resistance gene were tested. To assess the presence or absence of R Mh-chc A, the primer combinations eACAmCCT, eACTmCAT and eAGAmCTC were analysed, and for R Mh-tar , pAGmAAG, pAGmAGG (two marker loci) and pAGmAGA were used.
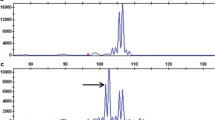
Of all seven markers tested, only three markers generated an informative polymorphic pattern diagnostic for the presence of the resistance locus: eACAmCCT_383 and eACTmCAT_165 for R Mh-chc A and pAGmAGA_221 for R Mh-tar (see Fig. 2). Marker pAGmAGA_221 was previously known as pAGmAGA_226 (Draaistra 2006). The distance of 17 cM between the marker and the R gene will somewhat decrease the diagnostic value of this marker because approx. 17% of the offspring may show recombination. The fragment mobility of 226 was obtained with a 33P-labelled Eco primer followed by autoradiography, whereas the mobility of 221 refers to the same fragment obtained with fluorescently labelled primers and separation on a LICOR system. The congruence of fragment mobilities between these two AFLP methods cannot be predicted in advance. One marker (pAGmAAG_111) could not be traced back, possibly due to methylation of the PstI site or a shift in mobility, where the fragment is hidden underneath other AFLP fragments.
Linkage groups indicating the position of AFLP markers and the position of R Mh-chc A and R Mh-tar (Draaistra 2006). The diagnostic value of the marker pattern in this pyramiding study is indicated with a plus or minus sign, where the sign indicates diagnostic or uninformative segregation pattern, respectively
Statistical Analysis
Statistical analyses were performed with MS-Excel and Genstat 10th Edition. The number of egg masses counted per root system were transformed by taking the natural logarithm of the (egg mass number + 1), to obtain a more uniform distribution of the variance. The heritability was estimated as follows: \( {h^2} = \sigma_{\text{g}}^2/\left( {\sigma_{\text{g}}^2 + \sigma_{\text{e}}^2/n} \right) \), where n is the number of replications, \( \sigma_{\text{e}}^2 \) is the expected mean square of the residuals and \( \sigma_{\text{g}}^2 \) is retrieved from the one-way ANOVA table as follows: \( E\left( {\text{MS}} \right){\text{genotypes}} = n\sigma_{\text{g}}^2 + \sigma_{\text{e}}^2 \). The individual and combined effects of R Mh-chc A and R Mh-tar were analysed with a general ANOVA. The size of the root system was visually classified according to an arbitrary scale of 1 to 5, and was taken as a covariate in ANOVA and a block structure was included in the model of: (repetition*genotype)/plant.
Results
Genotyping of Pyramiding Population RH04-456
The 148 offspring genotypes of the pyramiding population RH04-456 were genotyped with markers eACAmCCT_383 and eACTmCAT_165 to infer the presence of R Mh-chc A, while pAGmAGA_221 was used for R Mh-tar . Due to missing values and recombination events between markers eACAmCCT_383 and eACTmCAT_165, ten and 26 genotypes, respectively, were excluded from further analysis. In total, 112 genotypes remained for testing in the disease experiment. The pyramiding population could be differentiated into four groups: 30 without R genes, 27 with R Mh-tar present, 26 with R Mh-chc A present and 29 containing both R genes R Mh-tar + R Mh-chc A pyramided. The observed numbers of genotypes in each of the four groups complied with the expected 1:1:1:1 segregation ratio (χ 2 = 0.357; P = 0.948).
Meloidogyne hapla Disease Experiment
To test for resistance to M. hapla, 112 genotypes were inoculated with isolate “Bovensmilde”. The number of egg masses developed in the root system of individual plants of the pyramiding population differed between zero and 221. An example of egg masses stained red with Phloxine-B on a susceptible plant is shown in Fig. 3. The parental clones used as control displayed a high level of resistance. The Solanum chacoense × tuberosum hybrid 87-206-6 developed, on average, one egg mass per root system, the S. tuberosum × tarijense hybrid RH90-011-4, on average, three egg masses. The susceptible control cv. Nicola developed, on average, 127 egg masses. The numbers of egg masses were converted by a natural logarithm to reduce the variance before further data analysis. According to ANOVA, the genotypes showed significant differences in their number of egg masses (F ratio = 8.12; df = 530; P < 0.001). The heritability of the offspring population was 0.92.
The Individual and Joint Effect of the Loci R Mh-tar + R Mh-chc A on the Level of Resistance Against M. hapla
The size of the root system significantly influenced the number of egg masses (F ratio = 6.06; df = 1; P < 0.015), and was taken as a covariate in further analysis.
The pyramiding population, differentiated into four groups, based on flanking molecular markers, allowed estimating the effect of the loci R Mh-tar + R Mh-chc A. The group without R Mh-chc A and R Mh-tar developed, on average, 25.8 egg masses (see Table 1). Figure 4 shows the distribution of the mean number of egg masses per offspring genotype in four groups, which specify the absence or presence of the R genes. The group of genotypes containing R Mh-tar , but not R Mh-chc A, developed, on average, 11.8 egg masses, which means a decline of 55% in developed egg masses compared to the group without R Mh-chc A and R Mh-tar . However, according to ANOVA, this effect of R Mh-tar on the number of egg masses is not significant (F ratio = 1.02; df = 1; P = 0.315).
The group containing R Mh-chc A, but not R Mh-tar , developed, on average, 3.1 egg masses. The effect of R Mh-chc A is highly significant (F ratio = 23.29; df = 1; P < 0.001) and offers a reduction of 88% of the number of egg masses as compared to the group without R Mh-chc A and R Mh-tar .
When both R genes are present, the average number of egg masses is 3.0 (ln (egg mass + 1) = 0.76). Compared to the effect of 3.1 egg masses caused by R Mh-chc A alone, the joint effect of both loci, R Mh-tar and R Mh-chc A, does not further reduce the number of egg masses. This is also evident from the ANOVA, where an interaction between R Mh-tar and R Mh-chc A (F ratio = 3.06; df = 1; P = 0.083) was observed. Hence, the locus R Mh-chc A shows an epistatic interaction over the R Mh-tar locus.
Discussion
The objective of this research was to investigate if pyramiding of two M. hapla resistance genes allowed a further reduction of the number of egg masses as compared to the reduction obtained by the separate QTL for resistance. Due to the large variation in egg masses in the groups containing R Mh-tar , and the relatively small group sizes, the effect of R Mh-tar was not significant. The average decline of 55% in egg masses suggests, however, that the effect of R Mh-tar is probably real. Due to the lack of statistical significance of the contribution of R Mh-tar , we now can only speculate on the effect of pyramiding R Mh-chc A and R Mh-tar . The results of this experiment seem to suggest that pyramiding R Mh-chc A and R Mh-tar does not lead to an additional reduction in the number of egg masses, and hence, the level of resistance has not improved.
This study also provides a validation of the value of the R Mh-chc A locus. The R Mh-chc A locus was first identified as a QTL explaining 38% of the phenotypic variance and causing a 70% reduction of the number of egg masses, relative to susceptible offspring, in a BC1 mapping population (Draaistra 2006). In this study, the effect of the R Mh-chc A locus was even stronger, causing 88% reduction. Therefore, the value of this gene for potato breeding is beyond doubt, although cultivar registration authorities may require a higher level of reduction before a cultivar can be called resistant according to their criteria.
The R Mh-tar locus was initially identified in a BC1 mapping population, where a clear-cut Mendelian segregation of resistance was observed. Nevertheless, the R Mh-tar locus explained only 20% of the phenotypic variance in the mapping population, which demonstrates the great variability in the number of egg masses that can develop on an individual plant. This great variability also had a great impact on the statistical analysis of the QTL effects in this experiment. Although a clear 50% reduction of egg masses was observed, the effect was not significant. Hence, the validation of R Mh-tar was not achieved, but R Mh-tar probably remains a valuable gene for breeders. In retrospect, to obtain statistical significance for small-effect QTL, the offspring size of the experimental population should have been larger than the current 112 descendants, distributed over four classes with 26–30 plants per class.
In addition to the trait variability, the genetic distance of 17 cM between R Mh-tar and the diagnostic AFLP marker pAGmAGA_221 posed a second complication. Because of this genetic distance it is likely that some genotypes have recombination events between AFLP marker pAGmAGA_221 and resistance locus R Mh-tar , and thus, not containing the resistance locus, incorrectly entered the R Mh-tar group. The decline of 55% caused by R Mh-tar is therefore likely to be an underestimation of the actual effect. The same reasoning holds true, of course, for the groups without R Mh-tar . In retrospect, we should have used the AFLP markers that mapped closer to R Mh-tar , but of which the diagnostic allele was found in both parents of the pyramiding population. These markers were not tested, but could have resulted in a 3:1 segregating AFLP polymorphism, where marker presence would not be informative. The offspring of the pyramiding population without the diagnostic band would certainly not have R Mh-tar . Thus, with a marker that is heterozygous in both parents, a theoretic 25% of the offspring could still be genotyped. In this study, it could have eliminated 25% of the wrongly classified genotypes with a recombination event between the used AFLP marker pAGmAGA_221 and resistance locus R Mh-tar .
The heritability of 92% obtained in this study clearly demonstrates that the experimental error between plants within a genotype is very low. Hence, the resistance level of an individual genotype has been determined with high accuracy. The high variance between genotypes within R gene classes caused the lack of statistical significance of the effect of R Mh-tar . A striking observation is the discrepancy between the high heritability of this experiment with the modest explained variance of R Mh-chc A and R Mh-tar as observed in the initial mapping studies of Draaistra (2006). Usually, the gap between high heritability and low explained QTL variance is ascribed to genetic background. This could point to the putative significance of R Mh-chc B, also known to segregate in this population. More likely, however, this is another confirmation that recombination between the marker and R Mh-tar has compromised the statistical significance of this experiment.
In retrospect, the experimental design of this study should not have neglected the contribution of the R Mh-chc B locus, which descends from the resistant parent 87-206-6. This small-effect QTL explained only 9.6% of the phenotypic variance (Draaistra 2006), which was deemed insignificant and negligible. However, it might have had its own unique value in this pyramiding study. This unique value depends on the mutual complementarities of the R gene and the unique composition of each different M. hapla population.
The experiment may also have been compromised by the difference between the M. hapla nematode populations. Population Hi from the location “Smilde” was used to detect R Mh-chc A, and population Hb from the location “Zwaanshoek” was used to detect R Mh-tar (Draaistra 2006). These populations have been characterised by Van der Beek et al. (1998). In this pyramiding study, a different population from “Bovensmilde” was used, which was commercially available from RZ Research, Metslawier, The Netherlands. This issue demonstrates the interdependency of plant geneticists and nematologists and the necessity to maintain public accessible collections of well characterised research materials for a reasonable price.
The nature of the nematode population has great implications for the research of plant geneticists. At this moment, there is little information on the composition and allele frequency of avirulence genes within M. hapla populations. Differences between populations can greatly affect the identification of loci involved in nematode resistance and the estimates of the size of the QTL effect. A small-effect QTL such as R Mh-tar might appear as a large-effect QTL or as a single R gene with absolute effect, depending on the composition of the nematode population. The ease at which nematode populations can change their composition was demonstrated by Janssen et al. (1998), who could develop R gene breaking Meloidogyne chitwoodi populations in one generation, by selection of largely non-breaking isolates and subsequent multiplication on tomato. Furthermore, the efforts required to avoid admixture of M. hapla populations with individuals from M. chitwoodi and M. fallax are not trivial.
Genotyping the pyramiding population with the available AFLP markers proved to be problematic. Polymorphic patterns observed in a specific mapping population cannot always be expected in other populations. In this pyramiding population, the parents of both mapping populations were used to construct the pyramiding population to assure that the marker allele associated with the resistance locus will at least be recognised in the newly generated population. In this study, three cases were observed, where the marker alleles originally diagnostic for the disease in the original mapping study were also observed in the other parent, invalidating its diagnostic value. The diagnostic value of marker alleles across unrelated genotypes or different mapping populations than the mapping population in which the markers were developed can be improved if such an assay infers the haplotypes based on multiple SNPs (Sattarzadeh et al. 2006). In the study of Sattarzadeh et al. (2006), the allele-specific PCR primers were sensitive to SNPs that were diagnostic for a specific haplotype, but even this strategy resulted in a few false positive/false negative results in spite of the efforts to maximise the diagnostic value of the marker.
References
Dickson DW, Struble FB (1965) A sieving-staining technique for extraction of egg masses of Meloidogyne incognita from soil. Phytopathology 55:497
Draaistra J (2006) Genetic analysis of root-knot nematode resistance in potato. PhD thesis Laboratory of Plant Breeding. Wageningen University, The Netherlands. ISBN 90-8504-340-9. http://library.wur.nl/wda/dissertations/dis3880.pdf
Janssen GJW, Janssen R, van Norel A, Verkerk-Bakker B, Hoogendoorn J (1996a) Expression of resistance to the root-knot nematodes, Meloidogyne hapla and M. fallax, in wild Solanum spp. under field conditions. Eur J Plant Pathol 102:859–865
Janssen GJW, van Norel A, Verkerk-Bakker B, Janssen R (1996b) Resistance to Meloidogyne chitwoodi, M. fallax and M. hapla in wild tuber-bearing Solanum spp. Euphytica 92:287–294
Janssen GJW, Scholten OE, van Norel A, Hoogendoorn C (1998) Selection of virulence in Meloidogyne chitwoodi to resistance in the wild potato Solanum fendleri. Eur J Plant Pathol 104:645–651
Macguidwin AE, Rouse DI (1990) Effect of Meloidogyne hapla, alone and in combination with subthreshold populations of Verticillium dahliae, on disease symptomology and yield of potato. Phytopathology 80:482–486
Mugniéry D (2007) The canon of potato science: 15. Root-knot Nematodes. Potato Res 50(3–4):263–265
Sattarzadeh A, Achenbach U, Lubeck J, Strahwald J, Tacke E, Hofferbert HR, Rothsteyn T, Gebhardt C (2006) Single nucleotide polymorphism (SNP) genotyping as basis for developing a PCR-based marker highly diagnostic for potato varieties with high resistance to Globodera pallida pathotype Pa2/3. Mol Breeding 18:301–312
Stewart CN, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14:748–750
Van der Beek JG, Poleij LM, Zijlstra C, Janssen R, Janssen GJW (1998) Variation in virulence within Meloidogyne chitwoodi, M. fallax, and M. hapla on Solanum spp. Phytopathology 88:658–665
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Acknowledgements
We would like to acknowledge Robert Machado for technical assistance in the disease test, Petra van den Berg for laboratory assistance and Chris Maliepaard and Joao Paulo for their statistical advice.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Tan, M.Y.A., Alles, R., Hutten, R.C.B. et al. Pyramiding of Meloidogyne hapla Resistance Genes in Potato Does Not Result in an Increase of Resistance. Potato Res. 52, 331–340 (2009). https://doi.org/10.1007/s11540-009-9137-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11540-009-9137-2