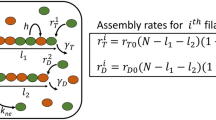
Abstract
The microtubule cytoskeleton is responsible for sustained, long-range intracellular transport of mRNAs, proteins, and organelles in neurons. Neuronal microtubules must be stable enough to ensure reliable transport, but they also undergo dynamic instability, as their plus and minus ends continuously switch between growth and shrinking. This process allows for continuous rebuilding of the cytoskeleton and for flexibility in injury settings. Motivated by in vivo experimental data on microtubule behavior in Drosophila neurons, we propose a mathematical model of dendritic microtubule dynamics, with a focus on understanding microtubule length, velocity, and state-duration distributions. We find that limitations on microtubule growth phases are needed for realistic dynamics, but the type of limiting mechanism leads to qualitatively different responses to plausible experimental perturbations. We therefore propose and investigate two minimally-complex length-limiting factors: limitation due to resource (tubulin) constraints and limitation due to catastrophe of large-length microtubules. We combine simulations of a detailed stochastic model with steady-state analysis of a mean-field ordinary differential equations model to map out qualitatively distinct parameter regimes. This provides a basis for predicting changes in microtubule dynamics, tubulin allocation, and the turnover rate of tubulin within microtubules in different experimental environments.








Similar content being viewed by others
Data availability
Experimental data is referenced appropriately throughout the manuscript and all simulation data is available upon request.
References
Akhmanova A, Kapitein LC (2022) Mechanisms of microtubule organization in differentiated animal cells. Nat Rev Mol Cell Biol 23(8):541–558
Alexandrova VV, Anisimov MN, Zaitsev AV et al (2022) Theory of tip structure-dependent microtubule catastrophes and damage-induced microtubule rescues. Proc Natl Acad Sci 119(46):e2208294119
Antal T, Krapivsky PL, Redner S et al (2007) Dynamics of an idealized model of microtubule growth and catastrophe. Phys Rev E Stat Nonlinear Soft Matter Phys. https://doi.org/10.1103/PhysRevE.76.041907
Baas PW, Lin S (2011) Hooks and comets: the story of microtubule polarity orientation in the neuron. Dev Neurobiol 71(6):403–418
Baas PW, Rao AN, Matamoros AJ et al (2016) Stability properties of neuronal microtubules. Cytoskeleton 73(9):442–460
Barlukova A, White D, Henry G et al (2018) Mathematical modeling of microtubule dynamic instability: new insight into the link between gtp-hydrolysis and microtubule aging. ESAIM Math Model Numer Anal 52(6):2433–2456
Bartolini F, Gundersen GG (2006) Generation of noncentrosomal microtubule arrays. J Cell Sci 119(20):4155–4163
Bolterauer H, Limbach HJ, Tuszyński J (1999) Models of assembly and disassembly of individual microtubules: stochastic and averaged equations. J Biol Phys 25:1–22
Bowne-Anderson H, Zanic M, Kauer M et al (2013) Microtubule dynamic instability: a new model with coupled gtp hydrolysis and multistep catastrophe. BioEssays 35(5):452–461
Buxton GA, Siedlak SL, Perry G et al (2010) Mathematical modeling of microtubule dynamics: insights into physiology and disease. Prog Neurobiol 92:478–483. https://doi.org/10.1016/j.pneurobio.2010.08.003
Cassimeris LU, Wadsworth P, Salmon E (1986) Dynamics of microtubule depolymerization in monocytes. J Cell Biol 102(6):2023–2032
Chen L, Stone MC, Tao J et al (2012) Axon injury and stress trigger a microtubule-based neuroprotective pathway. Proc Natl Acad Sci 109(29):11842–11847
Coombes CE, Yamamoto A, Kenzie MR et al (2013) Evolving tip structures can explain age-dependent microtubule catastrophe. Curr Biol 23(14):1342–1348
Deymier PA, Yang Y, Hoying J (2005) Effect of tubulin diffusion on polymerization of microtubules. Phys Rev E Stat Nonlinear Soft Matter Phys. https://doi.org/10.1103/PhysRevE.72.021906
Dogterom M, Leibler S (1993) Physical aspects of the growth and regulation of microtubule structures microtubules (mts) are long, rigid polymers made of tubulin-a globular protein found in eukaryotic cells [1]. Phys Rev Lett 70:1347–1350
Dogterom M, Maggs AC, Leibler S (1995) Diffusion and formation of microtubule asters: physical processes versus biochemical regulation. Proc Natl Acad Sci 92:6683–6688
Fees CP, Moore JK (2019) A unified model for microtubule rescue. Mol Biol Cell 30(6):753–765
Feng C, Thyagarajan P, Shorey M et al (2019) Patronin-mediated minus end growth is required for dendritic microtubule polarity. J Cell Biol 218:2309–2328. https://doi.org/10.1083/jcb.201810155
Flyvbjerg H, Holy TE, Leibler S (1996) Microtubule dynamics: caps, catastrophes, and coupled hydrolysis. Phys Rev E 54(5):5538
Gardner MK, Zanic M, Gell C et al (2011) Depolymerizing kinesins kip3 and mcak shape cellular microtubule architecture by differential control of catastrophe. Cell 147(5):1092–1103
Gardner MK, Zanic M, Howard J (2013) Microtubule catastrophe and rescue. Curr Opin Cell Biol 25(1):14–22
Gillespie DT (2001) Approximate accelerated stochastic simulation of chemically reacting systems. J Chem Phys 115(4):1716–1733
Gillespie DT et al (2007) Stochastic simulation of chemical kinetics. Annu Rev Phys Chem 58(1):35–55
Gliksman NR, Parsons SF, Salmon E (1992) Okadaic acid induces interphase to mitotic-like microtubule dynamic instability by inactivating rescue. J Cell Biol 119(5):1271–1276
Govindan BS, Spillman WB (2004) Steady states of a microtubule assembly in a confined geometry. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Top 70:4. https://doi.org/10.1103/PhysRevE.70.032901
Hertzler JI, Simonovitch SI, Albertson RM et al (2020) Kinetochore proteins suppress neuronal microtubule dynamics and promote dendrite regeneration. Mol Biol Cell 31(18):2125–2138
Hill TL (1984) Introductory analysis of the gtp-cap phase-change kinetics at the end of a microtubule (aggregation/polymer in solution/polymer on site/steady-state distribution). Proc Natl Acad Sci 81:6728–6732
Hinow P, Rezania V, Tuszyński JA (2009) Continuous model for microtubule dynamics with catastrophe, rescue, and nucleation processes. Phys Rev E 80(3):031904
Honoré S, Hubert F, Tournus M et al (2019) A growth-fragmentation approach for modeling microtubule dynamic instability. Bull Math Biol 81:722–758
Ji XY, Feng XQ (2011) Mechanochemical modeling of dynamic microtubule growth involving sheet-to-tube transition. PLoS ONE. https://doi.org/10.1371/journal.pone.0029049
Jonasson EM, Mauro AJ, Li C et al (2020) Behaviors of individual microtubules and microtubule populations relative to critical concentrations: dynamic instability occurs when critical concentrations are driven apart by nucleotide hydrolysis. Mol Biol Cell 31(7):589–618
Kapitein LC, Hoogenraad CC (2011) Which way to go? cytoskeletal organization and polarized transport in neurons. Mol Cell Neurosci 46(1):9–20
Kapitein LC, Hoogenraad CC (2015) Building the neuronal microtubule cytoskeleton. Neuron 87(3):492–506
Keating TJ, Borisy GG (1999) Centrosomal and non-centrosomal microtubules. Biol Cell 91(4–5):321–329
Kelliher MT, Saunders HA, Wildonger J (2019) Microtubule control of functional architecture in neurons. Curr Opin Neurobiol 57:39–45
Margolin G, Gregoretti IV, Goodson HV et al (2006) Analysis of a mesoscopic stochastic model of microtubule dynamic instability. Phys Rev E Stat Nonlinear Soft Matter Phys. https://doi.org/10.1103/PhysRevE.74.041920
Margolin G, Gregoretti IV, Cickovski TM et al (2012) The mechanisms of microtubule catastrophe and rescue: implications from analysis of a dimer-scale computational model. Mol Biol Cell 23(4):642–656
Margolis RL, Wilson L (1998) Microtubule treadmilling: what goes around comes around. BioEssays 20(10):830–836
Mazilu I, Zamora G, Gonzalez J (2010) A stochastic model for microtubule length dynamics. Physica A 389:419–427. https://doi.org/10.1016/j.physa.2009.10.017
Mitchison T, Kirschner M (1984) Dynamic instability of microtubule growth. Nature 312(5991):237–242
Molodtsov MI, Ermakova EA, Shnol EE et al (2005) A molecular-mechanical model of the microtubule. Biophys J 88(5):3167–3179
Nye DM, Albertson RM, Weiner AT et al (2020) The receptor tyrosine kinase ror is required for dendrite regeneration in drosophila neurons. PLoS Biol 18(3):e3000657
Padinhateeri R, Kolomeisky AB, Lacoste D (2012) Random hydrolysis controls the dynamic instability of microtubules. Biophys J 102:1274–1283. https://doi.org/10.1016/j.bpj.2011.12.059
Rank M, Mitra A, Reese L et al (2018) Limited resources induce bistability in microtubule length regulation. Phys Rev Lett. https://doi.org/10.1103/PhysRevLett.120.148101
Rodionov VI, Borisy GG (1997) Microtubule treadmilling in vivo. Science 275(5297):215–218
Rolls MM, Thyagarajan P, Feng C (2021) Microtubule dynamics in healthy and injured neurons. Dev Neurobiol 81:321–332. https://doi.org/10.1002/dneu.22746
Sanchez AD, Feldman JL (2017) Microtubule-organizing centers: from the centrosome to non-centrosomal sites. Curr Opin Cell Biol 44:93–101
Schulze E, Kirschner M (1986) Microtubule dynamics in interphase cells. J Cell Biol 102(3):1020–1031
Stone MC, Nguyen MM, Tao J et al (2010) Global up-regulation of microtubule dynamics and polarity reversal during regeneration of an axon from a dendrite. Mol Biol Cell 21:767–777. https://doi.org/10.1091/mbc.E09
Tao J, Feng C, Rolls MM (2016) The microtubule-severing protein fidgetin acts after dendrite injury to promote their degeneration. J Cell Sci 129:3274–3281. https://doi.org/10.1242/jcs.188540
Thyagarajan P, Feng C, Lee D et al (2022) Microtubule polarity is instructive for many aspects of neuronal polarity. Dev Biol 486:56–70
Varga V, Helenius J, Tanaka K et al (2006) Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol 8(9):957–962
Varga V, Leduc C, Bormuth V et al (2009) Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell 138:1174–1183. https://doi.org/10.1016/j.cell.2009.07.032
Verde F, Labbé JC, Dorée M et al (1990) Regulation of microtubule dynamics by cdc2 protein kinase in cell-free extracts of xenopus eggs. Nature 343(6255):233–238
White D, de Vries G, Martin J et al (2015) Microtubule patterning in the presence of moving motor proteins. J Theor Biol 382:81–90
Zakharov P, Gudimchuk N, Voevodin V et al (2015) Molecular and mechanical causes of microtubule catastrophe and aging. Biophys J 109:2574–2591. https://doi.org/10.1016/j.bpj.2015.10.048
Acknowledgements
This work was supported by NIH grant R01NS121245. ACN was partially supported by NSF grant DMS-2038056.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A: Stochastic Model Implementation Details
Appendix A: Stochastic Model Implementation Details
To implement the stochastic model of MT polymerization and depolymerization in Sect. 2, we implement an Euler-based stochastic scheme that is time- rather than event-driven, and thus we update events at fixed time steps \(\Delta t\). Similar to the leap condition in Gillespie’s tau-leaping scheme (Gillespie 2001; Gillespie et al. 2007), we choose a time interval \(\Delta t\) that is small enough so that reaction propensities are relatively fixed in time interval \([t_i, t_i+\Delta t = t_{i+1}]\), and large enough so that many reactions are still happening in each step. Since we are interested in the emergent behavior of dynamic MT instability, our approach differs from a strict implementation of tau-leaping in that we avoid resolving tubulin unit scale reactions and instead focus on MT end state switching reactions. Tau-leaping methods face significant challenges when one or more of the state variables are near boundary values. In our case, the pool of available tubulin is often near zero. This also motivates our choice of time scale, and our decision to treat tubulin resources as a continuum, rather that in discrete units. To determine how small time step \(\Delta t\) should be, we performed an experiment shown in Fig. 9 to see the effect of time step size on average length. We see MT behavior is qualitatively the same for time step sizes less than or equal to 1 s, so we utilize \(\Delta t = 1 \text { s}\).
To determine how long each end spends in a certain state during time step \(\Delta t\), we first update the length-dependent catastrophe rate based on the length of the microtubules at the previous time step, \(t_i\), denoted as \(L_i = L(t_i)\). Therefore, the catastrophe rate at \(t_{i+1} = t_i + \Delta t\) will take the following form
where \(\phi (x) = x - 1\). Again, for small L and large \(\gamma \), it is possible that \( \lambda _{min} > \lambda _{\textrm{g} \rightarrow \textrm{s}}^\pm + \gamma L_0(\frac{L(t)}{L_0} - 1)\), so then the catastrophe rate will be \(\lambda _{min}\) for short MTs. The updated catastrophe rates are then used to draw times spent in growth phase.
At each time step from \(t_i\) to \(t_i + \Delta t\), each microtubule end is in either a shrinking or growth state. For each end of the microtubule, the time spent in growth and shrinking, denoted as \(t_g\) and \(t_s\), respectively, are given by random variables drawn from the appropriate exponential distribution, where
We then determine if the microtubule exits the time step in either growth or shrinking based on the idea that only one shrinking phase can occur in a time step \(\Delta t\) (see Fig. 10). For example, if a microtubule end entered in growth and \(t_g > \Delta t\), then the microtubule would also exit the time step in growth (see Fig. 10d). Similarily, if a microtubule end entered in growth and \(t_g + t_s < \Delta t\), then the microtubule also exits in growth, shown in Fig. 10e.
Once the time spent in each state has been determined, we then perform the shrinking events, which involve updating the microtubule lengths and adding the lost tubulin to unavailable tubulin bin. If the shrinking events resulted in a completely-catastrophed microtubule (that is, a microtubule with length less than or equal to zero), we reseed the microtubule at its previous position with length zero and both ends in a growth state. This reseeding procedure ensures that the number of microtubules remains the same throughout the simulation.
After performing shrinking events, we update the available tubulin pool from the unavailable tubulin pool at rate \(\tau _{tub}\) and then implement growth events, which depend on the available tubulin pool. To allow for a smooth transition in growth speed as the available tubulin pool becomes very small, we model the dependence of the microtubule growth on the available pool using Michaelis–Menten kinetics. For example, suppose the desired amount of tubulin for growth for a single MT is \(v_{g} t_g\), where \(v_{g}\) is the growth velocity and \(t_g\) is the time spent in growth for that MT in \(\Delta t\). Since growth is dependent on tubulin availability, we update the desired growth length for the j-th MT to be
where F is the available tubulin amount and \(F_{1/2}\) is the same Michaelis–Menten constant from Eq. (18).
We implement the growth events for all MTs in \(\Delta t\) and decrease F based on the total desired polymerization length, \(\sum _{i=1}^N {\tilde{x}}_i\). If tubulin resources are limited, it is possible that the available tubulin amount is less than the total desired polymerization length, so that \(F < \sum _{i=1}^N {\tilde{x}}_i\). To account for this, we divide the available tubulin amount proportionally to each MT based on the desired polymerization length for that time step. Therefore, the final length grown for the j-th MT, \(x_j\), is
We then deplete the tubulin used to grow MTs from the available tubulin pool, F, so that
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nelson, A.C., Rolls, M.M., Ciocanel, MV. et al. Minimal Mechanisms of Microtubule Length Regulation in Living Cells. Bull Math Biol 86, 58 (2024). https://doi.org/10.1007/s11538-024-01279-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11538-024-01279-z