Abstract
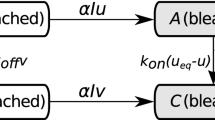
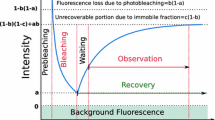
Identifying unique parameters for mathematical models describing biological data can be challenging and often impossible. Parameter identifiability for partial differential equations models in cell biology is especially difficult given that many established in vivo measurements of protein dynamics average out the spatial dimensions. Here, we are motivated by recent experiments on the binding dynamics of the RNA-binding protein PTBP3 in RNP granules of frog oocytes based on fluorescence recovery after photobleaching (FRAP) measurements. FRAP is a widely-used experimental technique for probing protein dynamics in living cells, and is often modeled using simple reaction-diffusion models of the protein dynamics. We show that current methods of structural and practical parameter identifiability provide limited insights into identifiability of kinetic parameters for these PDE models and spatially-averaged FRAP data. We thus propose a pipeline for assessing parameter identifiability and for learning parameter combinations based on re-parametrization and profile likelihoods analysis. We show that this method is able to recover parameter combinations for synthetic FRAP datasets and investigate its application to real experimental data.










Similar content being viewed by others
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Alexander AM, Lawley SD (2022) Inferences from FRAP data are model dependent: a subdiffusive analysis. Biophys J 121(20):3795–3810
Audoly S, Bellu G, D’Angio L, Saccomani MP, Cobelli C (2001) Global identifiability of nonlinear models of biological systems. IEEE Trans Biomed Eng 48(1):55–65
Browning AP, Tască M, Falcó C, Baker RE (2023) Structural identifiability analysis of linear reaction-advection-diffusion processes in mathematical biology. arXiv preprint arXiv:2309.15326
Cabral SE, Otis JP, Mowry KL (2022) Multivalent interactions with RNA drive recruitment and dynamics in biomolecular condensates in Xenopus oocytes. iScience 25:104811
Chis O-T, Banga JR, Balsa-Canto E (2011) Structural identifiability of systems biology models: a critical comparison of methods. PloS ONE 6(11):27755
Cintrón-Arias A, Banks HT, Capaldi A, Lloyd AL (2009) A sensitivity matrix based methodology for inverse problem formulation
Ciocanel M-V, Kreiling JA, Gagnon JA, Mowry KL, Sandstede B (2017) Analysis of active transport by fluorescence recovery after photobleaching. Biophys J 112(8):1714–1725
Cobelli C, Distefano JJ 3rd (1980) Parameter and structural identifiability concepts and ambiguities: a critical review and analysis. Am J Physiol-Regul Integr Comp Physiol 239(1):7–24
Cox SM, Matthews PC (2002) Exponential time differencing for stiff systems. J Comput Phys 176(2):430–455
Deschout H, Hagman J, Fransson S, Jonasson J, Rudemo M, Lorén N, Braeckmans K (2010) Straightforward FRAP for quantitative diffusion measurements with a laser scanning microscope. Opt Express 18(22):22886–22905
Eisenberg M (2019) Input-output equivalence and identifiability: some simple generalizations of the differential algebra approach. arXiv preprint arXiv:1302.5484
Eisenberg MC, Hayashi MA (2014) Determining identifiable parameter combinations using subset profiling. Math Biosci 256:116–126
Eisenberg MC, Robertson SL, Tien JH (2013) Identifiability and estimation of multiple transmission pathways in cholera and waterborne disease. J Theor Biol 324:84–102
Evans ND, Chappell MJ (2000) Extensions to a procedure for generating locally identifiable reparameterisations of unidentifiable systems. Math Biosci 168(2):137–159
Geweke JF et al (1991) Evaluating the accuracy of sampling-based approaches to the calculation of posterior moments. Technical report, Federal Reserve Bank of Minneapolis
GitHub: Sample Matlab code for estimating parameters from FRAP microscopy data. GitHub (2020)
Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR, Sethna JP (2007) Universally sloppy parameter sensitivities in systems biology models. PLoS Computat Biol 3(10):189
Haario H, Laine M, Mira A, Saksman E (2006) DRAM: efficient adaptive MCMC. Stat Comput 16(4):339–354
Hengl S, Kreutz C, Timmer J, Maiwald T (2007) Data-based identifiability analysis of non-linear dynamical models. Bioinformatics 23(19):2612–2618
Hines KE, Middendorf TR, Aldrich RW (2014) Determination of parameter identifiability in nonlinear biophysical models: a Bayesian approach. J General Physiol 143(3):401–416
Hong H, Ovchinnikov A, Pogudin G, Yap C (2020) Global identifiability of differential models. Commun Pure Appl Math 73(9):1831–1879
Ishikawa-Ankerhold HC, Ankerhold R, Drummen GP (2012) Advanced fluorescence microscopy techniques-FRAP, FLIP, FLAP, FRET and FLIM. Molecules 17(4):4047–4132
Kassam A-K, Trefethen LN (2005) Fourth-order time-stepping for stiff PDEs. SIAM J Sci Comput 26(4):1214–1233
Kenworthy AK (2023) What’s past is prologue: FRAP keeps delivering 50 years later. Biophys J 122:P3577-3586
Ljung L, Glad T (1994) On global identifiability for arbitrary model parametrizations. Automatica 30(2):265–276
Miao H, Xia X, Perelson AS, Wu H (2011) On identifiability of nonlinear ode models and applications in viral dynamics. SIAM Rev 53(1):3–39
Murphy SA, Van der Vaart AW (2000) On profile likelihood. J Am Stat Assoc 95(450):449–465
Neil CR, Jeschonek SP, Cabral SE, O’Connell LC, Powrie EA, Otis JP, Wood TR, Mowry KL (2021) L-bodies are RNA-protein condensates driving RNA localization in Xenopus oocytes. Mol Biol Cell 32(22):37
Ollivier F (1990) Le problème de l’identifiabilité structurelle globale: approche théorique, méthodes effectives et bornes de complexité. PhD thesis, Palaiseau, Ecole polytechnique
Powrie EA, Ciocanel V, Kreiling JA, Gagnon JA, Sandstede B, Mowry KL (2016) Using in vivo imaging to measure RNA mobility in Xenopus laevis oocytes. Methods 98:60–65
Raue A, Kreutz C, Maiwald T, Bachmann J, Schilling M, Klingmüller U, Timmer J (2009) Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics 25(15):1923–1929
Raue A, Kreutz C, Theis FJ, Timmer J (2013) Joining forces of Bayesian and frequentist methodology: a study for inference in the presence of non-identifiability. Philos Trans R Soc A Math Phys Eng Sci 371(1984):20110544
Reid J (1977) Structural identifiability in linear time-invariant systems. IEEE Trans Autom Control 22(2):242–246
Renardy M, Kirschner D, Eisenberg M (2022) Structural identifiability analysis of age-structured PDE epidemic models. J Math Biol 84(1):1–30
Siekmann I, Sneyd J, Crampin EJ (2012) MCMC can detect nonidentifiable models. Biophys J 103(11):2275–2286
Simpson MJ, Baker RE, Vittadello ST, Maclaren OJ (2020) Practical parameter identifiability for spatio-temporal models of cell invasion. J R Soc Interface 17(164):20200055
Sprague BL, Pego RL, Stavreva DA, McNally JG (2004) Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J 86(6):3473–3495
Acknowledgements
Ding, Mastromatteo, and Reichheld were partially supported by the NSF under grant DMS-174429. Sandstede was partially supported by the NSF under grants DMS-2038039 and DMS-2106566. The experimental work was funded by R01GM071049 from the NIH to Mowry.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A: Derivation of the Sensitivity Equations
Appendix A: Derivation of the Sensitivity Equations
We provide details on deriving the sensitivity equations associated with PDE model system (1). Here, we show the derivation of the PDE equations for the sensitivities with respect to the diffusion coefficient D, since the other sensitivities can be derived in a similar way.
We first consider the PDE for the free protein concentration:
We denote \(f_D=\frac{\partial f}{\partial D}\) and \(c_D=\frac{\partial c}{\partial D}\). We differentiate equation (29) with respect to the diffusion coefficient D and assume that the protein concentrations are smooth and thus have continuous partial derivatives. Applying the chain rule yields:
Similarly, consider the PDE for the bound protein concentration:
Differentiating this equation with respect to D yields the sensitivity equation:
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ciocanel, MV., Ding, L., Mastromatteo, L. et al. Parameter Identifiability in PDE Models of Fluorescence Recovery After Photobleaching. Bull Math Biol 86, 36 (2024). https://doi.org/10.1007/s11538-024-01266-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11538-024-01266-4