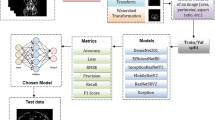
Abstract
Medical image classification plays a pivotal role within the field of medicine. Existing models predominantly rely on supervised learning methods, which necessitate large volumes of labeled data for effective training. However, acquiring and annotating medical image data is both an expensive and time-consuming endeavor. In contrast, semi-supervised learning methods offer a promising approach by harnessing limited labeled data alongside abundant unlabeled data to enhance the performance of medical image classification. Nonetheless, current methods often encounter confirmation bias due to noise inherent in self-generated pseudo-labels and the presence of boundary samples from different classes. To overcome these challenges, this study introduces a novel framework known as boundary sample-based class-weighted semi-supervised learning (BSCSSL) for medical image classification. Our method aims to alleviate the impact of intra- and inter-class boundary samples derived from unlabeled data. Specifically, we address reliable confidential data and inter-class boundary samples separately through the utilization of an inter-class boundary sample mining module. Additionally, we implement an intra-class boundary sample weighting mechanism to extract class-aware features specific to intra-class boundary samples. Rather than discarding such intra-class boundary samples outright, our approach acknowledges their intrinsic value despite the difficulty associated with accurate classification, as they contribute significantly to model prediction. Experimental results on widely recognized medical image datasets demonstrate the superiority of our proposed BSCSSL method over existing semi-supervised learning approaches. By enhancing the accuracy and robustness of medical image classification, our BSCSSL approach yields considerable implications for advancing medical diagnosis and future research endeavors.
Graphical abstract





Similar content being viewed by others
References
Moor M, Banerjee O, Abad ZSH, Krumholz HM, Leskovec J, Topol EJ, Rajpurkar P (2023) Foundation models for generalist medical artificial intelligence. Nature 616(7956):259–265
Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S, Dean J (2019) A guide to deep learning in healthcare. Nat Med 25(1):24–29
Suganyadevi S, Seethalakshmi V, Balasamy K (2022) A review on deep learning in medical image analysis. Int J Multimed Inf Retr 11(1):19–38
Eckardt J-N, Bornhäuser M, Wendt K, Middeke JM (2022) Semi-supervised learning in cancer diagnostics. Front Onco 12:960984
Yang X, Song Z, King I, Xu Z (2022) A survey on deep semi-supervised learning. IEEE Transactions on Knowledge and Data Engineering
Song Z, Yang X, Xu Z, King I (2022) Graph-based semi-supervised learning: a comprehensive review. IEEE Transactions on Neural Networks and Learning Systems
Zhang W, Zhu L, Hallinan J, Zhang S, Makmur A, Cai Q, Ooi BC (2022) BoostMIS: boosting medical image semi-supervised learning with adaptive pseudo labeling and informative active annotation. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 20666–20676
Luo Y, Liu W, Fang T, Song Q, Min X, Wang M, Li A (2023) CARL: cross-aligned representation learning for multi-view lung cancer histology classification. In: Greenspan H, Madabhushi A, Mousavi P, Salcudean S, Duncan J, Syeda-Mahmood T, Taylor R (eds) Medical image computing and computer assisted intervention - MICCAI 2023. Springer Nature Switzerland, Cham, pp 358–367
Atban F, Ekinci E, Garip Z (2023) Traditional machine learning algorithms for breast cancer image classification with optimized deep features. Biomed Signal Process Control 81:104534
Yu J-G, Wu Z, Ming Y, Deng S, Wu Q, Xiong Z, Yu T, Xia G-S, Jiang Q, Li Y (2023) Bayesian collaborative learning for whole-slide image classification. IEEE Transactions on Medical Imaging
Ling Y, Wang Y, Dai W, Yu J, Liang P, Kong D (2023) MTANet: multi-task attention network for automatic medical image segmentation and classification. IEEE Transactions on Medical Imaging
Yang Y, Fu H, Aviles-Rivero AI, Schönlieb C-B, Zhu L (2023) DiffMIC: dual-guidance diffusion network for medical image classification. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 95–105
Zeng Q, Xie Y, Lu Z, Xia Y (2023) PEFAT: boosting semi-supervised medical image classification via pseudo-loss estimation and feature adversarial training. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 15671–15680
Gao Z, Hong B, Li Y, Zhang X, Wu J, Wang C, Zhang X, Gong T, Zheng Y, Meng D et al (2023) A semi-supervised multi-task learning framework for cancer classification with weak annotation in whole-slide images. Med Image Anal 83:102652
Yang Q, Chen Z, Yuan Y (2023) Hierarchical bias mitigation for semi-supervised medical image classification. IEEE Transactions on Medical Imaging
Wang J, Qiao L, Zhou S, Zhou J, Wang J, Li J, Ying S, Chang C, Shi J (2024) Weakly supervised lesion detection and diagnosis for breast cancers with partially annotated ultrasound images. IEEE Transactions on Medical Imaging
Peng Z, Tian S, Yu L, Zhang D, Wu W, Zhou S (2023) Semi-supervised medical image classification with adaptive threshold pseudo-labeling and unreliable sample contrastive loss. IEEE Trans Med Imaging 79:104142
Chen X, Yu G, Tan Q, Wang J (2019) Weighted samples based semi-supervised classification. Appl Soft Comput 79:46–58
Camargo G, Bugatti PH, Saito PT (2020) Active semi-supervised learning for biological data classification. PLoS One 15(8):e0237428
Aromal M, Rasool A, Dubey A, Roy B (2021) Optimized weighted samples based semi-supervised learning. In: 2021 Second international conference on electronics and sustainable communication systems (ICESC), IEEE, pp 1311–1318
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
B. J. A. Kingma DP (2014) Adam: a method for stochastic optimization. In: Internetional conferrence for learning representations
Zhu W, Liu J, Huang Y (2023) HNSSL: hard negative-based self-supervised learning. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 4777–4786
Tarvainen A, Valpola H (2017) Mean teachers are better role models: weight-averaged consistency targets improve semi-supervised deep learning results. Advances in Neural Information Processing Systems 30
Sohn K, Berthelot D, Carlini N, Zhang Z, Zhang H, Raffel CA, Cubuk ED, Kurakin A, Li C-L (2020) FixMatch: simplifying semi-supervised learning with consistency and confidence. Adv Neural Inf Process Syst 33:596–608
Z. Hu, Z. Yang, X. Hu, R. Nevatia, (2021) Simple: similar pseudo label exploitation for semi-supervised classification. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 15099–15108
Li J, Xiong C, Hoi SC (2021) CoMatch: semi-supervised learning with contrastive graph regularization. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 9475–9484
Zheng M, You S, Huang L, Wang F, Qian C, Xu C (2022) SimMatch: semi-supervised learning with similarity matching. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 14471–14481
Liu Q, Yu L, Luo L, Dou Q, Heng PA (2020) Semi-supervised medical image classification with relation-driven self-ensembling model. IEEE Trans Med Imaging 39(11):3429–3440
Guo L-Z, Zhang Z-Y, Jiang Y, Li Y-F, Zhou Z-H (2020) Safe deep semi-supervised learning for unseen-class unlabeled data. In: International conference on machine learning. PMLR, pp 3897–3906
Hang W, Huang Y, Liang S, Lei B, Choi K-S, Qin J (2022) Reliability-aware contrastive self-ensembling for semi-supervised medical image classification. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 754–763
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fang, P., Feng, R., Liu, C. et al. Boundary sample-based class-weighted semi-supervised learning for malignant tumor classification of medical imaging. Med Biol Eng Comput (2024). https://doi.org/10.1007/s11517-024-03114-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11517-024-03114-y