Abstract
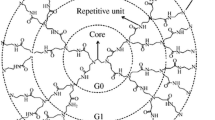
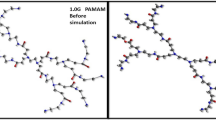
Poly(amidoamine) (PAMAM) dendrimers are widely studied as drug vectors due to their particular structure and excellent properties. Drug molecules can be loaded in the internal cavity of dendrimers or adsorbed on the surface. In this paper, the interaction force and spatial configuration between PAMAM and several typical chemotherapy drugs (doxorubicin (DOX), paclitaxel (TAX), hydroxycamptothecin (HCPT)) is studied by molecular dynamics (MD) simulations. Several essential parameters of dendrimers as drug carriers are analyzed by combining experiments and MD simulations, including particle size, drug loading, drug release, and biocompatibility. The simulation of dendrimer@drug complexes demonstrates that many cavities are semi-open, and most drug molecules are not completely wrapped. The internal structure of PAMAM dendrimers became looser with more extended nuclei, which increased the non-covalent interactions between PAMAM dendrimers and drug molecules. The umbrella sampling simulations reveal that the change of binding energies between dendrimers and drug molecules is responsible for the variation in drug release rate. This study provides valuable enlightenment information for the drug loading/release of PAMAM dendrimers.
Similar content being viewed by others
References
Lu Y, Slomberg D L, Shah A, et al. Nitric oxide-releasing amphiphilic poly(amidoamine) (PAMAM) dendrimers as antibacterial agents. Biomacromolecules, 2013, 14: 3589–3598
Zhou L, Shan Y, Hu H, et al. Synthesis and biomedical applications of dendrimers. Curr Org Chem, 2018, 22: 600–612
Cong H, Zhou L, Meng Q, et al. Preparation and evaluation of PA-MAM dendrimer-based polymer gels physically cross-linked by hydrogen bonding. Biomater Sci, 2019, 7: 3918–3925
Maji S, Agarwal T, Maiti T K. PAMAM (generation 4) incorporated gelatin 3D matrix as an improved dermal substitute for skin tissue engineering. Colloids Surfs B-Biointerfaces, 2017, 155: 128–134
Luong D, Kesharwani P, Deshmukh R, et al. PEGylated PAMAM dendrimers: Enhancing efficacy and mitigating toxicity for effective anticancer drug and gene delivery. Acta Biomater, 2016, 43: 14–29
Sun Y, Ma X, Jing X, et al. PAMAM-functionalized cellulose nano-crystals with needle-like morphology for effective cancer treatment. Nanomaterials, 2021, 11: 1640
Sonuç Karaboğa M N, Sezgintürk M K. Determination of C-reactive protein by PAMAM decorated ITO based disposable biosensing system: A new immunosensor design from an old molecule. Talanta, 2018, 186: 162–168
Sun Y, Hu H, Jing X, et al. Co-delivery of chemotherapeutic drugs and cell cycle regulatory agents using nanocarriers for cancer therapy. Sci China Mater, 2021, 64: 1827–1848
Sun Y, Jing X, Ma X, et al. Versatile types of polysaccharide-based drug delivery systems: From strategic design to cancer therapy. Int J Mol Sci, 2020, 21: 9159
Sun Q, Sun X, Ma X, et al. Integration of nanoassembly functions for an effective delivery cascade for cancer drugs. Adv Mater, 2014, 26: 7615–7621
Xiong Z, Shen M, Shi X. Dendrimer-based strategies for cancer therapy: Recent advances and future perspectives. Sci China Mater, 2018, 61: 1387–1403
Zou W T, Zhong W, Sun M G, et al. pH-Responsive dendrimer-functionalized cotton cellulose nanocrystals for effective cancer treatment. Ferroelectrics, 2021, 578: 108–112
Wu J S, Li J X, Shu N, et al. A polyamidoamine (PAMAM) derivative dendrimer with high loading capacity of TLR7/8 agonist for improved cancer immunotherapy. Nano Res, 2022, 15: 510–518
Pishavar E, Oroojalian F, Salmasi Z, et al. Recent advances of dendrimer in targeted delivery of drugs and genes to stem cells as cellular vehicles. Biotechnol Prog, 2021, 37: e3174
Ren L, Lv J, Wang H, et al. A coordinative dendrimer achieves excellent efficiency in cytosolic protein and peptide delivery. Angew Chem Int Ed, 2020, 59: 4711–4719
Zhang C, Pan D, Luo K, et al. Dendrimer-doxorubicin conjugate as enzyme-sensitive and polymeric nanoscale drug delivery vehicle for ovarian cancer therapy. Polym Chem, 2014, 5: 5227–5235
Tian W, Ma Y. Theoretical and computational studies of dendrimers as delivery vectors. Chem Soc Rev, 2013, 42: 705–727
Sathe R Y, Bharatam P V. Drug-dendrimer complexes and conjugates: Detailed furtherance through theory and experiments. Adv Colloid Interface Sci, 2022, 303: 102639
Tian F, Lin X, Valle R P, et al. Poly(amidoamine) dendrimer as a respiratory nanocarrier: Insights from experiments and molecular dynamics simulations. Langmuir, 2019, 35: 5364–5371
Wang B, Sun Y, Davis T P, et al. Understanding effects of PAMAM dendrimer size and surface chemistry on serum protein binding with discrete molecular dynamics simulations. ACS Sustain Chem Eng, 2018, 6: 11704–11715
Liu Y, Bryantsev V S, Diallo M S, et al. PAMAM dendrimers undergo pH responsive conformational changes without swelling. J Am Chem Soc, 2009, 131: 2798–2799
Jafari M, Mehrnejad F, Talandashti R, et al. Effect of the lipid composition and cholesterol on the membrane selectivity of low generations PAMAM dendrimers: A molecular dynamics simulation study. Appl Surf Sci, 2021, 540: 148274
Maingi V, Kumar M V S, Maiti P K. PAMAM dendrimer-drug interactions: Effect of pH on the binding and release pattern. J Phys Chem B, 2012, 116: 4370–4376
Jain V, Maiti P K, Bharatam P V. Atomic level insights into realistic molecular models of dendrimer-drug complexes through MD simulations. J Chem Phys, 2016, 145: 124902
Jing X, Sun Y, Liu Y, et al. Alginate/chitosan-based hydrogel loaded with gene vectors to deliver polydeoxyribonucleotide for effective wound healing. Biomater Sci, 2021, 9: 5533–5541
Raemdonck K, Demeester J, de Smedt S. Advanced nanogel engineering for drug delivery. Soft Matter, 2009, 5: 707–715
Van Der Spoel D, Lindahl E, Hess B, et al. GROMACS: Fast, flexible, and free. J Comput Chem, 2005, 26: 1701–1718
Martínez L, Andrade R, Birgin E G, et al. PACKMOL: A package for building initial configurations for molecular dynamics simulations. J Comput Chem, 2009, 30: 2157–2164
Kästner J. Umbrella sampling. WIREs Comput Mol Sci, 2011, 1: 932–942
Gao W, Jiao Y, Dai L L. The effects of size, shape, and surface composition on the diffusive behaviors of nanoparticles at/across water-oil interfaces via molecular dynamics simulations. J Nanopart Res, 2016, 18: 91
Barra P A, Barraza L, Jiménez V A, et al. Complexation of mefenamic acid by low-generation PAMAM dendrimers: Insight from NMR spectroscopy studies and molecular dynamics simulations. Macromol Chem Phys, 2014, 215: 372–383
Hu J, Xu T, Cheng Y. NMR insights into dendrimer-based host-guest systems. Chem Rev, 2012, 112: 3856–3891
Meng Q, Hu H, Zhou L, et al. Logical design and application of prodrug platforms. Polym Chem, 2019, 10: 306–324
Marzinek J K, Bond P J, Lian G, et al. Free energy predictions of ligand binding to an α-helix using steered molecular dynamics and umbrella sampling simulations. J Chem Inf Model, 2014, 54: 2093–2104
Author information
Authors and Affiliations
Corresponding authors
Additional information
This work was supported by the National Natural Science Foundation of China (Grant Nos. 21874078, 22074072, and 22274083), the Taishan Young Scholar Program of Shandong Province (Grant No. tsqn20161027), the China Postdoctoral Science Foundation (Grant No. 2018M630752), the Postdoctoral Scientific Research Foundation of Qingdao, the Innovation and Development Joint Fund of Natural Science Foundation of Shandong Province (Grant No. ZR2022LZY022), the Science and Technology Planing Project of South District of Qingdao City (Grant No. 2022-4-005-YY), the Exploration Project of the State Key Laboratory of BioFibers and Eco-Textiles of Qingdao University (Grant No. TSKT202101), and the High Level Discipline Project of Shandong Province.
Supporting Information
The supporting information is available online at tech.scichina.com and link.springer.com. The supporting materials are published as submitted, without typesetting or editing. The responsibility for scientific accuracy and content remains entirely with the authors.
Rights and permissions
About this article
Cite this article
Zhou, L., Li, J., Yu, B. et al. The drug loading behavior of PAMAM dendrimer: Insights from experimental and simulation study. Sci. China Technol. Sci. 66, 1129–1140 (2023). https://doi.org/10.1007/s11431-022-2178-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11431-022-2178-8