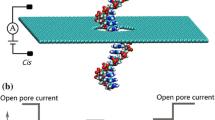
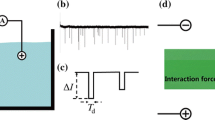
Abstract
Nanopore devices have attracted a lot of attention for their potential application in DNA sequencing. Here, we study how an occluding object placed near a nanopore affects its access resistance by integrating an atomic force microscopy with a nanopore sensor. It is found that there exists a critical hemisphere around the nanopore, inside which the tip of an atomic force microscopy will affect the ionic current. The radius of this hemisphere, which is a bit smaller than the theoretical capture radius of ions, increases linearly with the applied bias voltage and quadratically with the nanopore diameter, but is independent of the operation modes and scanning speeds of the atomic force microscopy. A theoretical model is also proposed to describe how the tip position and geometrical parameters affect the access resistance.
Similar content being viewed by others
References
Li J L, Gershow M, Stein D, et al. DNA molecules and configurations in a solid-state nanopore microscope. Nat Mater, 2003, 2: 611–615
Sha J J, Si W, Xu W, et al. Glass capillary nanopore for single molecule detection. Sci China Tech Sci, 2015, 58: 803–812
Wu G S, Zhang Y, Si W, et al. Integrated solid-state nanopore devices for third generation DNA sequencing. Sci China Tech Sci, 2014, 57: 1925–1935
Plesa C, van Loo N, Ketterer P, et al. Velocity of DNA during translocation through a solid state nanopore. Nano Lett, 2014, 15: 732–737
Nelson E M, Li H, Timp G. Direct, concurrent measurements of the forces and currents affecting DNA in a nanopore with comparable topography. Acs Nano, 2014, 8: 5484–5493
Zhang Y, Wu G, Ma J, et al. Temperature effect on translocation speed and capture rate of nanopore-based DNA detection. Sci China Tech Sci, 2014, 58: 519–525
Cracknell J A, Japrung D, Bayley H. Translocating kilobase rna through the staphylococcal α-hemolysin nanopore. Nano Lett, 2013, 13: 2500–2505
Clamer M, Höfler L, Mikhailova E, et al. Detection of 3'-end rna uridylation with a protein nanopore. Acs Nano, 2013, 8: 1364–1374
Steinbock L J, Krishnan S, Bulushev R D, et al. Probing the size of proteins with glass nanopores. Nanoscale, 2014, 6: 14380
Sha J, Hasan T, Milana S, et al. Nanotubes complexed with DNA and proteins for resistive-pulse sensing. Acs Nano, 2013, 7: 8857–8869
Liu S, Zhao Y, Parks J W, et al. Correlated electrical and optical analysis of single nanoparticles and biomolecules on a nanoporegated optofluidic chip. Nano Lett, 2014, 14: 4816–4820
German S R, Luo L, White H S, et al. Controlling nanoparticle dynamics in conical nanopores. J Phys Chem C, 2013, 117: 703–711
Si W, Sha J J, Liu L, et al. Effect of nanopore size on poly(dt)(30) translocation through silicon nitride membrane. Sci China Tech Sci, 2013, 56: 2398–2402
Si W, Zhang Y, Wu G S, et al. DNA sequencing technology based on nanopore sensors by theoretical calculations and simulations. Chin Sci Bull, 2014, 59: 4929–4941
Zwolak M, Di Ventra M. Colloquium: Physical approaches to DNA sequencing and detection. Rev Mod Phys, 2008, 80: 141–165
Shankla M, Aksimentiev A. Conformational transitions and stop-andgo nanopore transport of single-stranded DNA on charged graphene. Nat Commun, 2014, 5: 5171
Gopfrich K, Kulkarni C V, Pambos O J, et al. Lipid nanobilayers to host biological nanopores for DNA translocations. Langmuir, 2013, 29: 355–364
Akeson M, Branton D, Kasianowicz J J, et al. Microsecond timescale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single rna molecules. Biophys J, 1999, 77: 3227–3233
Maglia G, Restrepo M R, Mikhailova E, et al. Enhanced translocation of single DNA molecules through alpha-hemolysin nanopores by manipulation of internal charge. Proc Natl Acad Sci USA, 2008, 105: 19720–19725
Hoogerheide D P, Lu B, Golovchenko J A. Pressure-voltage trap for DNA near a solid-state nanopore. Acs Nano, 2014, 8: 7384–7391
Aksimentiev A, Heng J B, Timp G, et al. Microscopic kinetics of DNA translocation through synthetic nanopores. Biophys J, 2004, 87: 2086–2097
Binquan L. Numerically testing phenomenological models for conductance of a solid-state nanopore. Nanotechnology, 2015, 26: 055502
Kowalczyk S W, Grosberg A Y, Rabin Y, et al. Modeling the conductance and DNA blockade of solid-state nanopores. Nanotechnology, 2011, 22: 315101
Venkatesan B M, Bashir R. Nanopore sensors for nucleic acid analysis. Nat Nano, 2011, 6: 615–624
Xie P, Xiong Q H, Fang Y, et al. Local electrical potential detection of DNA by nanowire-nanopore sensors. Nat Nanotech, 2012, 7: 119–125
Kowalczyk S W, Dekker C. Measurement of the docking time of a DNA molecule onto a solid-state nanopore. Nano Lett, 2012, 12: 4159–4163
Keyser U F, Koeleman B N, Van Dorp S, et al. Direct force measurements on DNA in a solid-state nanopore. Nat Phys, 2006, 2: 473–477
Gollnick B, Carrasco C, Zuttion F, et al. Probing DNA helicase kinetics with temperature-controlled magnetic tweezers. Small, 2015, 11: 1273–1284
Hyun C, Kaur H, Rollings R, et al. Threading immobilized DNA molecules through a solid-state nanopore at >100 mus per base rate. Acs Nano, 2013, 7: 5892–5900
Zhang X, Liu B W, Servos M R, et al. Polarity control for nonthiolated DNA adsorption onto gold nanoparticles. Langmuir, 2013, 29: 6091–6098
Peng H, Ling X S. Reverse DNA translocation through a solid-state nanopore by magnetic tweezers. Nanotechnology, 2009, 20: 185101
Keyser U F, Krapf D, Koeleman B N, et al. Nanopore tomography of a laser focus. Nano Lett, 2005, 5: 2253–2256
Chen C C, Derylo M A, Baker L A. Measurement of ion currents through porous membranes with scanning ion conductance microscopy. Anal Chem, 2009, 81: 4742–4751
Hyun C, Rollings R, Li J L. Probing access resistance of solid-state nanopores with a scanning-probe microscope tip. Small, 2012, 8: 385–392
King G M, Golovchenko J A. Probing nanotube-nanopore interactions. Phys Rev Lett, 2005, 95: 216103
Hall J E. Access resistance of a small circular pore. J Gen Physiol, 1975, 66: 531–532
Lulevich V, Kim S, Grigoropoulos C P, et al. Frictionless sliding of single-stranded DNA in a carbon nanotube pore observed by single molecule force spectroscopy. Nano Lett, 2011, 11: 1171–1176
Wanunu M, Morrison W, Rabin Y, et al. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat Nanotechnol, 2010, 5: 160–165
Duan C, Majumdar A. Anomalous ion transport in 2-nm hydrophilic nanochannels. Nat Nano, 2010, 5: 848–852
Liu Q, Wu H, Wu L, et al. Voltage-driven translocation of DNA through a high throughput conical solid-state nanopore. Plos One, 2012, 7: e46014
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Si, W., Yang, H., Li, K. et al. Investigation on the interaction length and access resistance of a nanopore with an atomic force microscopy. Sci. China Technol. Sci. 60, 552–560 (2017). https://doi.org/10.1007/s11431-016-0494-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11431-016-0494-7