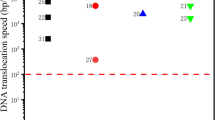
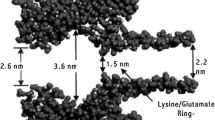
Abstract
DNA sequencing based on nanopore sensors is a promising tool for third-generation sequencing technology because of its special properties, such as revolutionized speed and low cost. With about two decades of nanopore technology development, the pioneering work has demonstrated the ability of nanopores to perform single-molecule detection and DNA sequencing. However, the microscopic mechanisms of DNA transport dynamics through nanopores remain largely unknown. Currently, DNA microscopic transport in a nanopore is difficult to characterize and several unexpected experimental observations are equivocal. This limitation can be resolved using theoretical calculations and simulations. These computational methods can monitor the entire dynamic process that DNA undergoes in solution at a single-atom resolution that can accurately unveil the mystery of DNA transport dynamics and predict certain unexpected phenomena. This paper mainly reports the recent applications of computational and simulation methods applied to the study of DNA transport through both biological and synthetic nanopores. We hope the theoretical calculations and simulations of DNA transport through nanopores can benefit the design of DNA sequencing devices.







Similar content being viewed by others
References
Mereuta L, Roy M, Asandei A et al (2014) Slowing down single-molecule trafficking through a protein nanopore reveals intermediates for peptide translocation. Sci Rep 4:3885
Ying YL, Zhang JJ, Meng FN et al (2013) A stimuli-responsive nanopore based on a photoresponsive host-guest system. Sci Rep 3:1662
Liu K, Feng J, Kis A et al (2014) Atomically thin molybdenum disulfide nanopores with high sensitivity for DNA translocation. ACS Nano 8:2504–2511
Zhou Z, Hu Y, Wang H et al (2013) DNA translocation through hydrophilic nanopore in hexagonal boron nitride. Sci Rep 3:3287
Chen YS, Lee CH, Hung MY et al (2013) DNA sequencing using electrical conductance measurements of a DNA polymerase. Nat Nantechnol 8:452–458
Schneider GF, Xu Q, Hage S et al (2013) Tailoring the hydrophobicity of graphene for its use as nanopores for DNA translocation. Nat Commun 4:2619
Liu S, Lu B, Zhao Q et al (2013) Boron nitride nanopores: highly sensitive DNA single-molecule detectors. Adv Mater 25:4549–4554
Li W, Bell NAW, Hernandez-Ainsa S et al (2013) Single protein molecule detection by glass nanopores. ACS Nano 7:4129–4134
Kasianowicz JJ, Brandin E, Branton D et al (1996) Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA 93:13770–13773
Venta K, Shemer G, Puster M et al (2013) Differentiation of short, single-stranded DNA homopolymers in solid-state nanopores. ACS Nano 7:4629–4636
Wen S, Zeng T, Liu L et al (2011) Highly sensitive and selective DNA-based detection of mercury(ii) with alpha-hemolysin nanopore. J Am Chem Soc 133:18312–18317
Yang C, Liu L, Zeng T et al (2013) Highly sensitive simultaneous detection of lead(ii) and barium(ii) with g-quadruplex DNA in α-hemolysin nanopore. Anal Chem 85:7302–7307
Keyser UF, Koeleman BN, Van Dorp S et al (2006) Direct force measurements on DNA in a solid-state nanopore. Nat Phys 2:473–477
Clarke J, Wu HC, Jayasinghe L et al (2009) Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol 4:265–270
Akeson M, Branton D, Kasianowicz JJ et al (1999) Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single rna molecules. Biophys J 77:3227–3233
Zwolak M, Di Ventra M (2008) Colloquium: physical approaches to DNA sequencing and detection. Rev Mod Phys 80:141–165
Purnell RF, Mehta KK, Schmidt JJ (2008) Nucleotide identification and orientation discrimination of DNA homopolymers immobilized in a protein nanopore. Nano Lett 8:3029–3034
Stoddart D, Heron AJ, Mikhailova E et al (2009) Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad Sci USA 106:7702–7707
Aksimentiev A, Schulten K (2005) Imaging alpha-hemolysin with molecular dynamics: ionic conductance, osmotic permeability, and the electrostatic potential map. Biophys J 88:3745–3761
Bhattacharya S, Derrington IM, Pavlenok M et al (2012) Molecular dynamics study of mspa arginine mutants predicts slow DNA translocations and ion current blockades indicative of DNA sequence. ACS Nano 6:6960–6968
Qiu H, Guo WL (2012) Detecting ssdna at single-nucleotide resolution by sub-2-nanometer pore in monoatomic graphene: A molecular dynamics study. Appl Phys Lett 100:083106
Aksimentiev A (2010) Deciphering ionic current signatures of DNA transport through a nanopore. Nanoscale 2:468–483
Venkatesan BM, Bashir R (2011) Nanopore sensors for nucleic acid analysis. Nat Nanotechnol 6:615–624
Branton D, Deamer DW, Marziali A et al (2008) The potential and challenges of nanopore sequencing. Nat Biotechnol 26:1146–1153
Min SK, Kim WY, Cho Y et al (2011) Fast DNA sequencing with a graphene-based nanochannel device. Nat Nanotechnol 6:162–165
Li J, Zhang Y, Yang J et al (2013) Molecular dynamics study of DNA translocation through graphene nanopores. Phys Rev E 87:062707
Lansac Y, Maiti PK, Glaser MA (2004) Coarse-grained simulation of polymer translocation through an artificial nanopore. Polymer 45:3099–3110
Tian P, Smith GD (2003) Translocation of a polymer chain across a nanopore: a brownian dynamics simulation study. J Chem Phys 119:11475–11483
Chen CM (2005) Driven translocation dynamics of polynucleotides through a nanopore: off-lattice monte-carlo simulations. Phys A 350:95–107
Lebrun A, Lavery R (1996) Modelling extreme stretching of DNA. Nucleic Acids Res 24:2260–2267
Maffeo C, Schopflin R, Brutzer H et al (2010) DNA-DNA interactions in tight supercoils are described by a small effective charge density. Phys Rev Lett 105:158101
Luan B, Aksimentiev A (2008) DNA attraction in monovalent and divalent electrolytes. J Am Chem Soc 130:15754–15755
Kong Y, Cui DX, Ozkan CS et al (2003) Modelling carbon nanotube based bio-nano systems: a molecular dynamics study. In: Ozkan CS, Santini JT, Gao H et al (eds) Symposium on biomicroelectromechanical systems (BioMEMS) held at the 2003 MRS Spring Meeting, San Francisco, April 2003. Materials research society symposium proceedings, vol 773. Materials Research Society, WARRENDALE, pp 111–116
Li Z, Yang W (2010) Capture and manipulation of hybrid dnas by carbon nanotube bundles. Nanotechnology 21:195301
Enyashin AN, Gemming S, Seifert G (2007) DNA-wrapped carbon nanotubes. Nanotechnology 18:245702
Liu YL, Iqbal SM (2009) A mesoscale model of DNA interaction with functionalized nanopore. Appl Phys Lett 95:223701
Wanunu M, Morrison W, Rabin Y et al (2010) Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat Nanotechnol 5:160–165
Hatlo MM, Panja D, van Roij R (2011) Translocation of DNA molecules through nanopores with salt gradients: the role of osmotic flow. Phys Rev Lett 107:068101
Wells DB, Abramkina V, Aksimentiev A (2007) Exploring transmembrane transport through alpha-hemolysin with grid-steered molecular dynamics. J Chem Phys 127:125101
Butler TZ, Pavlenok M, Derrington IM et al (2008) Single-molecule DNA detection with an engineered Mspa protein nanopore. Proc Natl Acad Sci USA 105:20647–20652
Derrington IM, Butler TZ, Collins MD et al (2010) Nanopore DNA sequencing with Mspa. Proc Natl Acad Sci USA 107:16060–16065
Aksimentiev A, Heng JB, Timp G et al (2004) Microscopic kinetics of DNA translocation through synthetic nanopores. Biophys J 87:2086–2097
Comer J, Dimitrov V, Zhao Q et al (2009) Microscopic mechanics of hairpin DNA translocation through synthetic nanopores. Biophys J 96:593–608
Schneider GF, Kowalczyk SW, Calado VE et al (2010) DNA translocation through graphene nanopores. Nano Lett 10:3163–3167
Siwy ZS, Davenport M (2010) Nanopores graphene opens up to DNA. Nat Nanotechnol 5:697–698
TraversiF RaillonC, Benameur SM et al (2013) Detecting the translocation of DNA through a nanopore using graphene nanoribbons. Nat Nanotechnol 8:939–945
Garaj S, Hubbard W, Reina A et al (2010) Graphene as a subnanometre trans-electrode membrane. Nature 467:190–193
Wells DB, Belkin M, Comer J et al (2012) Assessing graphene nanopores for sequencing DNA. Nano Lett 12:4117–4123
Sathe C, Zou X, Leburton JP et al (2011) Computational investigation of DNA detection using graphene nanopores. ACS Nano 5:8842–8851
Liu JW (2012) Adsorption of DNA onto gold nanoparticles and graphene oxide: surface science and applications. Phys Chem Chem Phys 14:10485–10496
Zhang X, Servos MR, Liu JW (2012) Surface science of DNA adsorption onto citrate-capped gold nanoparticles. Langmuir 28:3896–3902
Zhang X, Liu BW, Servos MR et al (2013) Polarity control for nonthiolated DNA adsorption onto gold nanoparticles. Langmuir 29:6091–6098
He H, Scheicher RH, Pandey R et al (2008) Functionalized nanopore-embedded electrodes for rapid DNA sequencing. J Phys Chem C 112:3456–3459
Nelson T, Zhang B, Prezhdo OV (2010) Detection of nucleic acids with graphene nanopores: ab initio characterization of a novel sequencing device. Nano Lett 10:3237–3242
Postma HWC (2010) Rapid sequencing of individual DNA molecules in graphene nanogaps. Nano Lett 10:420–425
Wilson J, Di Ventra M (2013) Single-base DNA discrimination via transverse ionic transport. Nanotechnology 24:415101
Shi XH, Yong K, Zhao YP et al (2005) Molecular dynamics simulation of peeling a DNA molecule on substrate. Acta Mech Sin 21:249–256
Neuman KC, Nagy A (2008) Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Meth 5:491–505
Spiering A, Getfert S, Sischka A et al (2011) Nanopore translocation dynamics of a single DNA-bound protein. Nano Lett 11:2978–2982
Lulevich V, Kim S, Grigoropoulos CP et al (2011) Frictionless sliding of single-stranded DNA in a carbon nanotube pore observed by single molecule force spectroscopy. Nano Lett 11:1171–1176
Zhang Z, Shen J, Wang H et al (2014) Effects of graphene nanopore geometry on DNA sequencing. J Phys Chem Lett 5:1602–1607
Liu Q, Wu H, Wu L et al (2012) Voltage-driven translocation of DNA through a high throughput conical solid-state nanopore. PLoS One 7:e46014
Meller A, Nivon L, Brandin E et al (2000) Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci USA 97:1079–1084
Kowalczyk SW, Wells DB, Aksimentiev A et al (2012) Slowing down DNA translocation through a nanopore in lithium chloride. Nano Lett 12:1038–1044
Zhang Y, Liu L, Sha J et al (2013) Nanopore detection of DNA molecules in magnesium chloride solutions. Nanoscale Res Lett 8:245
Fologea D, Uplinger J, Thomas B et al (2005) Slowing DNA translocation in a solid-state nanopore. Nano Lett 5:1734–1737
Si W, Sha J, Liu L et al (2013) Effect of nanopore size on poly(dt)30 translocation through silicon nitride membrane. Sci China Technol Sci 56:2398–2402
Utkur M, Jeffrey C, Valentin D et al (2010) Slowing the translocation of double-stranded DNA using a nanopore smaller than the double helix. Nanotechnology 21:395501
Lu B, Hoogerheide DP, Zhao Q et al (2013) Pressure-controlled motion of single polymers through solid-state nanopores. Nano Lett 13:3048–3052
Zhang H, Zhao Q, Tang Z et al (2013) Slowing down DNA translocation through solid-state nanopores by pressure. Small 9:4112–4117
Manrao EA, Derrington IM, Laszlo AH et al (2012) Reading DNA at single-nucleotide resolution with a mutant mspa nanopore and Phi29 DNA polymerase. Nat Biotechnol 30:349–353
Cherf GM, Lieberman KR, Rashid H et al (2012) Automated forward and reverse ratcheting of DNA in a nanopore at 5-Angstrom precision. Nat Biotechnol 30:344–348
Olasagasti F, Lieberman KR, Benner S et al (2010) Replication of individual DNA molecules under electronic control using a protein nanopore. Nat Nanotechnol 5:798–806
Lieberman KR, Cherf GM, Doody MJ et al (2010) Processive replication of single DNA molecules in a nanopore catalyzed by Phi29 DNA polymerase. J Am Chem Soc 132:17961–17972
Hyun C, Kaur H, Rollings R et al (2013) Threading immobilized DNA molecules through a solid-state nanopore at >100 Μs per base rate. ACS Nano 7:5892–5900
Luan B, Aksimentiev A (2010) Control and reversal of the electrophoretic force on DNA in a charged nanopore. J Phys Condens Matter 22:454123
Sigalov G, Comer J, Timp G et al (2008) Detection of DNA sequences using an alternating electric field in a nanopore capacitor. Nano Lett 8:56–63
Belkin M, Maffeo C, Wells DB et al (2013) Stretching and controlled motion of single-stranded DNA in locally heated solid-state nanopores. ACS Nano 7:6816–6824
Avdoshenko SM, Nozaki D, Gomes da Rocha C et al (2013) Dynamic and electronic transport properties of DNA translocation through graphene nanopores. Nano Lett 13:1969–1976
Luan BQ, Peng HB, Polonsky S et al (2010) Base-by-base ratcheting of single stranded DNA through a solid-state nanopore. Phys Rev Lett 104:238103
Acknowledgements
This work was supported by the National Basic Research Program of China (2011CB707605) and the National Natural Science Foundation of China (50925519, 51375092).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
SPECIAL TOPIC: Nanopore Analysis
About this article
Cite this article
Si, W., Zhang, Y., Wu, G. et al. DNA sequencing technology based on nanopore sensors by theoretical calculations and simulations. Chin. Sci. Bull. 59, 4929–4941 (2014). https://doi.org/10.1007/s11434-014-0622-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-014-0622-x