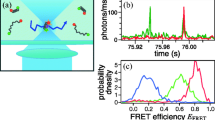
Abstract
Detecting protein-protein interactions (PPIs) provides fundamental information for understanding biochemical processes such as the transduction of signals from one cellular location to another; however, traditional biochemical techniques cannot provide sufficient spatio-temporal information to elucidate these molecular interactions in living cells. Over the past decade, several new techniques have enabled the identification and characterization of PPIs. In this review, we summarize three main techniques for detecting PPIs in vivo, focusing on their basic principles and applications in biological studies. We place a special emphasis on their advantages and limitations, and, in particular, we introduced some uncommon new techniques, such as single-molecule FRET (smFRET), FRET-fluorescence lifetime imaging microscopy (FRET-FLIM), cytoskeleton-based assay for protein-protein interaction (CAPPI) and single-molecule protein proximity index (smPPI), highlighting recent improvements to the established techniques. We hope that this review will provide a valuable reference to enable researchers to select the most appropriate technique for detecting PPIs.
Similar content being viewed by others
References
Abrahamsson, S., Chen, J., Hajj, B., Stallinga, S., Katsov, A.Y., Wisniewski, J., Mizuguchi, G., Soule, P., Mueller, F., Dugast Darzacq, C., et al. (2013). Fast multicolor 3D imaging using aberration-corrected multifocus microscopy. Nat Methods 10, 60–63.
Banaszynski, L.A., Liu, C.W., and Wandless, T.J. (2005). Characterization of the FKBP·rapamycin·FRB ternary complex. J Am Chem Soc 127, 4715–4721.
Bellati, J., Champeyroux, C., Hem, S., Rofidal, V., Krouk, G., Maurel, C., and Santoni, V. (2016). Novel aquaporin regulatory mechanisms revealed by interactomics. Mol Cell Proteomics 15, 3473–3487.
Bolte, S., and Cordelières, F.P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224, 213–232.
Bon, P., Linarès-Loyez, J., Feyeux, M., Alessandri, K., Lounis, B., Nassoy, P., and Cognet, L. (2018). Self-interference 3D super-resolution microscopy for deep tissue investigations. Nat Methods 15, 449–454.
Boute, N., Jockers, R., and Issad, T. (2002). The use of resonance energy transfer in high-throughput screening: BRET versus FRET. Trends Pharmacol Sci 23, 351–354.
Branchini, B. (2016). Firefly luciferase-based sequential bioluminescence resonance energy transfer (BRET)-fluorescence resonance energy transfer (FRET) protease assays. Methods Mol Biol 1461, 101–115.
Branchini, B.R., Rosenberg, J.C., Ablamsky, D.M., Taylor, K.P., Southworth, T.L., and Linder, S.J. (2011). Sequential bioluminescence resonance energy transfer—fluorescence resonance energy transferbased ratiometric protease assays with fusion proteins of firefly luciferase and red fluorescent protein. Anal Biochem 414, 239–245.
Bucherl, C., Aker, J., de Vries, S., and Borst, J.W. (2010). Probing proteinprotein interactions with FRET-FLIM. Methods Mol Biol 655, 389–399.
Bucherl, C.A., Bader, A., Westphal, A.H., Laptenok, S.P., and Borst, J.W. (2014). FRET-FLIM applications in plant systems. Protoplasma 251, 383–394.
Bucherl, C.A., Jarsch, I.K., Schudoma, C., Segonzac, C., Mbengue, M., Robatzek, S., MacLean, D., Ott, T., and Zipfel, C. (2017). Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. Elife 6, 1–28.
Carriba, P., Navarro, G., Ciruela, F., Ferré, S., Casadó, V., Agnati, L., Cortés, A., Mallol, J., Fuxe, K., Canela, E.I., et al. (2008). Detection of heteromerization of more than two proteins by sequential BRET-FRET. Nat Methods 5, 727–733.
Chang, C.W., and Kumar, S. (2013). Vinculin tension distributions of individual stress fibers within cell-matrix adhesions. J Cell Sci 126, 3021–3030.
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., Tang, X., and Zhou, J.M. (2008a). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146, 368–376.
Chen, P.Y., Deane, C.M., and Reinert, G. (2008b). Predicting and validating protein interactions using network structure. PLoS Comput Biol 4, e1000118.
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nürnberger, T., Jones, J.D.G., Felix, G., and Boller, T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500.
Ciruela, F. (2008). Fluorescence-based methods in the study of proteinprotein interactions in living cells. Curr Opin Biotech 19, 338–343.
Comeau, J.W.D., Kolin, D.L., and Wiseman, P.W. (2008). Accurate measurements of protein interactions in cells via improved spatial image cross-correlation spectroscopy. Mol Biosyst 4, 672–685.
Conrad, N.K. (2008). Co-immunoprecipitation techniques for assessing RNA-protein interactions in vivo. Method Enzymol 449, 317–342.
Conway, D.E., Breckenridge, M.T., Hinde, E., Gratton, E., Chen, C.S., and Schwartz, M.A. (2013). Fluid shear stress on endothelial cells modulates mechanical tension across VE-cadherin and PECAM-1. Curr Biol 23, 1024–1030.
Costes, S.V., Daelemans, D., Cho, E.H., Dobbin, Z., Pavlakis, G., and Lockett, S. (2004). Automatic and quantitative measurement of proteinprotein colocalization in live cells. Biophys J 86, 3993–4003.
Cui, Y., Li, X., Yu, M., Li, R., Fan, L., Zhu, Y., and Lin, J. (2018a). Sterols regulate endocytic pathways during flg22-induced defense responses in Arabidopsis. Development 145, dev165688.
Cui, Y., Yu, M., Yao, X., Xing, J., Lin, J., and Li, X. (2018b). Singleparticle tracking for the quantification of membrane protein dynamics in living plant cells. Mol Plant 11, 1315–1327.
Daniels, G.J., Jenkins, R.D., Bradshaw, D.S., and Andrews, D.L. (2003). Resonance energy transfer: The unified theory revisited. J Chem Phys 119, 2264–2274.
De Los Santos, C., Chang, C.W., Mycek, M.A., and Cardullo, R.A. (2015). FRAP, FLIM, and FRET: Detection and analysis of cellular dynamics on a molecular scale using fluorescence microscopy. Mol Reprod Dev 82, 587–604.
De Munter, S., Görnemann, J., Derua, R., Lesage, B., Qian, J., Heroes, E., Waelkens, E., Van Eynde, A., Beullens, M., and Bollen, M. (2017). Split-BioID: a proximity biotinylation assay for dimerization-dependent protein interactions. FEBS Lett 591, 415–424.
Demir, F., Horntrich, C., Blachutzik, J.O., Scherzer, S., Reinders, Y., Kierszniowska, S., Schulze, W.X., Harms, G.S., Hedrich, R., Geiger, D., et al. (2013). Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc Natl Acad Sci USA 110, 8296–8301.
Dragulescu-Andrasi, A., Chan, C.T., De, A., Massoud, T.F., and Gambhir, S.S. (2011). Bioluminescence resonance energy transfer (BRET) imaging of protein-protein interactions within deep tissues of living subjects. Proc Natl Acad Sci USA 108, 12060–12065.
Dunn, K.W., Kamocka, M.M., and McDonald, J.H. (2011). A practical guide to evaluating colocalization in biological microscopy. Am J Physiol-Cell Physiol 300, C723–C742.
Elangovan, M., Day, R.N., and Periasamy, A. (2002). Nanosecond fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy to localize the protein interactions in a single living cell. J Microsc 205, 3–14.
Fan, L., Hao, H., Xue, Y., Zhang, L., Song, K., Ding, Z., Botella, M.A., Wang, H., and Lin, J. (2013). Dynamic analysis of Arabidopsis AP2 subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development 140, 3826–3837.
Fan, M., Wang, M., and Bai, M.Y. (2016). Diverse roles of SERK family genes in plant growth, development and defense response. Sci China Life Sci 59, 889–896.
Forster T. (1948). Intermolecular energy migration and fluorescence. Ann Phys 2, 55–75.
Filonov, G.S., and Verkhusha, V.V. (2013). A near-infrared BiFC reporter for in vivo imaging of protein-protein interactions. Chem Biol 20, 1078–1086.
Fujii, Y., Yoshimura, A., and Kodama, Y. (2018). A novel orange-colored bimolecular fluorescence complementation (BiFC) assay using monomeric Kusabira-Orange protein. Biotechniques 64, 153–161.
Fujikawa, Y., and Kato, N. (2007). Technical advance: Split luciferase complementation assay to study protein-protein interactions in Arabidopsis protoplasts. Plant J 52, 185–195.
Gandia, J., Galino, J., Amaral, O.B., Soriano, A., Lluís, C., Franco, R., and Ciruela, F. (2008). Detection of higher-order G protein-coupled receptor oligomers by a combined BRET-BiFC technique. FEBS Lett 582, 2979–2984.
Gilles, J.F., Dos Santos, M., Boudier, T., Bolte, S., and Heck, N. (2017). DiAna, an ImageJ tool for object-based 3D co-localization and distance analysis. Methods 115, 55–64.
Ghosh, I., Hamilton, A.D., and Regan, L. (2000). Antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein. J Am Chem Soc 122, 5658–5659.
Gregorio, G.G., Masureel, M., Hilger, D., Terry, D.S., Juette, M., Zhao, H., Zhou, Z., Perez-Aguilar, J.M., Hauge, M., Mathiasen, S., et al. (2017). Single-molecule analysis of ligand efficacy in ß2AR–G-protein activation. Nature 547, 68–73.
Gustafsson, M.G.L., Shao, L., Carlton, P.M., Wang, C.J.R., Golubovskaya, I.N., Cande, W.Z., Agard, D.A., and Sedat, J.W. (2008). Threedimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J 94, 4957–4970.
Hajj, B., Wisniewski, J., El Beheiry, M., Chen, J., Revyakin, A., Wu, C., and Dahan, M. (2014). Whole-cell, multicolor superresolution imaging using volumetric multifocus microscopy. Proc Natl Acad Sci USA 111, 17480–17485.
Hayes, S., Malacrida, B., Kiely, M., and Kiely, P.A. (2016). Studying protein-protein interactions: progress, pitfalls and solutions. Biochem Soc Trans 44, 994–1004.
Helmuth, J.A., Paul, G., and Sbalzarini, I.F. (2010). Beyond colocalization: inferring spatial interactions between sub-cellular structures from microscopy images. BMC Bioinf 11, 372.
Hinterdorfer, P., and Van Oijen, A. (2009). Handbook of Single-molecule Biophysics (New York: Springer), pp. 43–93.
Huang, X., Fan, J., Li, L., Liu, H., Wu, R., Wu, Y., Wei, L., Mao, H., Lal, A., Xi, P., et al. (2018). Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat Biotechnol 36, 451–459.
Ishikawa-Ankerhold, H.C., Ankerhold, R., and Drummen, G.P.C. (2012). Advanced fluorescence microscopy techniques—FRAP, FLIP, FLAP, FRET and FLIM. Molecules 17, 4047–4132.
Jaqaman, K., Loerke, D., Mettlen, M., Kuwata, H., Grinstein, S., Schmid, S.L., and Danuser, G. (2008). Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods 5, 695–702.
Kim, D.I., Birendra, K.C., Zhu, W., Motamedchaboki, K., Doye, V., and Roux, K.J. (2014). Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci USA 111, E2453–E2461.
Kim, S.B., Ozawa, T., Watanabe, S., and Umezawa, Y. (2004). Highthroughput sensing and noninvasive imaging of protein nuclear transport by using reconstitution of split Renilla luciferase. Proc Natl Acad Sci USA 101, 11542–11547.
Kodama, Y., and Hu, C.D. (2012). Bimolecular fluorescence complementation (BiFC): A 5-year update and future perspectives. Biotechniques 53, 285–298.
Komis, G., Šamajová, O., Ovecka, M., and Šamaj, J. (2015). Superresolution microscopy in plant cell imaging. Trends Plant Sci 20, 834–843.
Kurihara, M., Ohmuro-Matsuyama, Y., Ayabe, K., Yamashita, T., Yamaji, H., and Ueda, H. (2016). Ultra sensitive firefly luciferase-based proteinprotein interaction assay (FlimPIA) attained by hinge region engineering and optimized reaction conditions. Biotech J 11, 91–99.
Lagache, T., Grassart, A., Dallongeville, S., Faklaris, O., Sauvonnet, N., Dufour, A., Danglot, L., and Olivo-Marin, J.C. (2018). Mapping molecular assemblies with fluorescence microscopy and object-based spatial statistics. Nat Commun 9, 698.
Lalonde, S., Ehrhardt, D.W., Loqué, D., Chen, J., Rhee, S.Y., and Frommer, W.B. (2008). Molecular and cellular approaches for the detection of protein-protein interactions: latest techniques and current limitations. Plant J 53, 610–635.
Li, L., Yu, Y., Zhou, Z., and Zhou, J.M. (2016). Plant pattern-recognition receptors controlling innate immunity. Sci China Life Sci 59, 878–888.
Li, Q. (2004). A syntaxin 1, Galpha(O), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J Neurosci 24, 4070–4081.
Li, S., Armstrong, C.M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P.O., Han, J.D., Chesneau, A., Hao, T., et al. (2004). A map of the interactome network of the metazoan C. elegans. Science 303, 540–543.
Li, X., Wang, X., Yang, Y., Li, R., He, Q., Fang, X., Luu, D.T., Maurel, C., and Lin, J. (2011). Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 23, 3780–3797.
Lin, J.S., and Lai, E.M. (2017). Protein-protein interactions: co-immunoprecipitation. Methods Mol Biol 1615, 211–219.
Liu, Z., Lavis, L.D., and Betzig, E. (2015). Imaging live-cell dynamics and structure at the single-molecule level. Mol Cell 58, 644–659.
Long, Y., Stahl, Y., Weidtkamp-Peters, S., Smet, W., Du, Y., Gadella Jr, T. W.J., Goedhart, J., Scheres, B., and Blilou, I. (2018). Optimizing FRETFLIM labeling conditions to detect nuclear protein interactions at native expression levels in living Arabidopsis roots. Front Plant Sci 9.
Luker, K.E., Smith, M.C.P., Luker, G.D., Gammon, S.T., Piwnica-Worms, H., and Piwnica-Worms, D. (2004). Kinetics of regulated proteinprotein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci USA 101, 12288–12293.
Lv, S., Miao, H., Luo, M., Li, Y., Wang, Q., Julie Lee, Y.R., and Liu, B. (2017). CAPPI: a cytoskeleton-based localization assay reports proteinprotein interaction in living cells by fluorescence microscopy. Mol Plant 10, 1473–1476.
Ma, Y., Wang, Y., Zhou, X., Kuang, C., and Liu, X. (2014). 3D dual-virtualpinhole assisted single particle tracking microscopy. J Opt 16, 075703.
Maizel, A., von Wangenheim, D., Federici, F., Haseloff, J., and Stelzer, E. H.K. (2011). High-resolution live imaging of plant growth in near physiological bright conditions using light sheet fluorescence microscopy. Plant J 68, 377–385.
McDonald, J.H., and Dunn, K.W. (2014). Statistical tests for measures of colocalization in biological microscopy. J Microsc 252, 295–302.
Miller, K.E., Kim, Y., Huh, W.K., and Park, H.O. (2015). Bimolecular fluorescence complementation (BiFC) analysis: advances and recent applications for genome-wide interaction studies. J Mol Biol 427, 2039–2055.
Ovecka, M., von Wangenheim, D., Tomancák, P., Šamajová, O., Komis, G., and Šamaj, J. (2018). Multiscale imaging of plant development by lightsheet fluorescence microscopy. Nat Plants 4, 639–650.
Ozawa, T. (2009). Protein reconstitution methods for visualizing biomolecular function in living cells. Yakugaku Zasshi 129, 289–295.
Periasamy, A., and Day, R.N. (1999). Visualizing protein interactions in living cells using digitized GFP imaging and FRET microscopy. Method Cell Biol 58, 293–314.
Perillo, E.P., Liu, Y.L., Ang, P., Dunn, A.K., and Yeh, H.C. (2017). A 3D dual-particle tracking co-localization microscope for the study of dna dynamics in free solution. Biophys J 112, 298a.
Perillo, E.P., Liu, Y.L., Huynh, K., Liu, C., Chou, C.K., Hung, M.C., Yeh, H.C., and Dunn, A.K. (2015). Deep and high-resolution threedimensional tracking of single particles using nonlinear and multiplexed illumination. Nat Commun 6, 1–12.
Pfleger, K.D.G., and Eidne, K.A. (2006). Illuminating insights into proteinprotein interactions using bioluminescence resonance energy transfer (BRET). Nat Meth 3, 165–174.
Pi, J., Jin, H., Yang, F., Chen, Z.W., and Cai, J. (2014). In situ single molecule imaging of cell membranes: linking basic nanotechniques to cell biology, immunology and medicine. Nanoscale 6, 12229–12249.
Reichmann, D., Rahat, O., Cohen, M., Neuvirth, H., and Schreiber, G. (2007). The molecular architecture of protein-protein binding sites. Curr Opin Struct Biol 17, 67–76.
Remy, I., and Michnick, S.W. (2007). Application of protein-fragment complementation assays in cell biology. Biotechniques 42, 137–145.
Sambrook, J., and Russell, D.W. (2006). Detection of protein-protein interactions using the GST fusion protein pulldown technique. CSH Protocols 1, 275–276.
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682.
Schopp, I.M., Amaya Ramirez, C.C., Debeljak, J., Kreibich, E., Skribbe, M., Wild, K., and Béthune, J. (2017). Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat Commun 8, 15690.
Schuler, B., and Eaton, W.A. (2008). Protein folding studied by singlemolecule FRET. Curr Opin Struct Biol 18, 16–26.
Shaw, S.L., and Ehrhardt, D.W. (2013). Smaller, faster, brighter: advances in optical imaging of living plant cells. Annu Rev Plant Biol 64, 351–375.
Shrestha, D., Jenei, A., Nagy, P., Vereb, G., and Szöllosi, J. (2015). Understanding FRET as a research tool for cellular studies. IJMS 16, 6718–6756.
Shyu, Y.J., Suarez, C.D., and Hu, C.D. (2008). Visualization of ternary complexes in living cells by using a BiFC-based FRET assay. Nat Protoc 3, 1693–1702.
Stirnweiss, A., Hartig, R., Gieseler, S., Lindquist, J.A., Reichardt, P., Philipsen, L., Simeoni, L., Poltorak, M., Merten, C., Zuschratter, W., et al. (2013). T cell activation results in conformational changes in the Src family kinase Lck to induce its activation. Sci Signal 6, ra13.
Subramanian, C., Woo, J., Cai, X., Xu, X., Servick, S., Johnson, C.H., Nebenführ, A., and von Arnim, A.G. (2006). A suite of tools and application notes for in vivo protein interaction assays using bioluminescence resonance energy transfer (BRET). Plant J 48, 138–152.
Sun, S.H., Yang, X.B., Wang, Y., and Shen, X.H. (2016). In vivo analysis of protein-protein interactions with bioluminescence resonance energy transfer (BRET): progress and prospects. Int J Mol Sci 17, 1–21.
Suzuki, A., Long, S.K., and Salmon, E.D. (2018). An optimized method for 3D fluorescence co-localization applied to human kinetochore protein architecture. eLife 7, e32418.
Vogelsang, J., Doose, S., Sauer, M., and Tinnefeld, P. (2007). Singlemolecule fluorescence resonance energy transfer in nanopipets: improving distance resolution and concentration range. Anal Chem 79, 7367–7375.
Wang, L., Li, H., Lv, X., Chen, T., Li, R., Xue, Y., Jiang, J., Jin, B., Baluška, F., Šamaj, J., et al. (2015). Spatiotemporal dynamics of the BRI1 receptor and its regulation by membrane microdomains in living Arabidopsis cells. Mol Plant 8, 1334–1349.
Wang, L., Xue, Y., Xing, J., Song, K., and Lin, J. (2018). Exploring the spatiotemporal organization of membrane proteins in living plant cells. Annu Rev Plant Biol 69, 525–551.
Wu, Y., Eghbali, M., Ou, J., Lu, R., Toro, L., and Stefani, E. (2010). Quantitative determination of spatial protein-protein correlations in fluorescence confocal microscopy. Biophys J 98, 493–504.
Xia, Z., and Rao, J. (2009). Biosensing and imaging based on bioluminescence resonance energy transfer. Curr Opin Biotech 20, 37–44.
Xie, Q., Soutto, M., Xu, X., Zhang, Y., and Johnson, C.H. (2011). Bioluminescence resonance energy transfer (BRET) imaging in plant seedlings and mammalian cells. Method Mol Biol 680, 3–28.
Xu, X., Soutto, M., Xie, Q., Servick, S., Subramanian, C., von Arnim, A. G., and Hirschie Johnson, C. (2007). Imaging protein interactions with bioluminescence resonance energy transfer (BRET) in plant and mammalian cells and tissues. Proc Natl Acad Sci USA 104, 10264–10269.
Xue, Y., Xing, J., Wan, Y., Lv, X., Fan, L., Zhang, Y., Song, K., Wang, L., Wang, X., Deng, X., et al. (2018). Arabidopsis blue light receptor phototropin 1 undergoes blue light-induced activation in membrane microdomains. Mol Plant 11, 846–859.
Zhou, J.M., and Yang, W.C. (2016). Receptor-like kinases take center stage in plant biology. Sci China Life Sci 59, 863–866.
Zhu, Y., Wang, Y., Li, R., Song, X., Wang, Q., Huang, S., Jin, J.B., Liu, C. M., and Lin, J. (2010). Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J 61, 223–233.
Zinchuk, V., Wu, Y., and Grossenbacher-Zinchuk, O. (2013). Bridging the gap between qualitative and quantitative colocalization results in fluorescence microscopy studies. Sci Rep 3, 1–5.
Zinchuk, V., Wu, Y., Grossenbacher-Zinchuk, O., and Stefani, E. (2011). Quantifying spatial correlations of fluorescent markers using enhanced background reduction with protein proximity index and correlation coefficient estimations. Nat Protoc 6, 1554–1567.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31530084, 31761133009) and the Programme of Introducing Talents of Discipline to Universities (111 project, B13007).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cui, Y., Zhang, X., Yu, M. et al. Techniques for detecting protein-protein interactions in living cells: principles, limitations, and recent progress. Sci. China Life Sci. 62, 619–632 (2019). https://doi.org/10.1007/s11427-018-9500-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-018-9500-7