Abstract
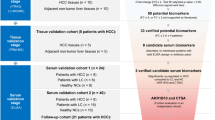
For successful therapy, hepatocellular carcinoma (HCC) must be detected at an early stage. Herein, we used a proteomic approach to analyze the secretory/releasing proteome of HCC tissues to identify plasma biomarkers. Serum-free conditioned media (CM) were collected from primary cultures of cancerous tissues and surrounding noncancerous tissues. Proteomic analysis of the CM proteins permitted the identification of 1365 proteins. The enriched molecular functions and biological processes of the CM proteins, such as hydrolase activity and catabolic processes, were consistent with the liver being the most important metabolic organ. Moreover, 19% of the proteins were characterized as extracellular or membrane-bound. For validation, secretory proteins involved in transforming growth factor-β signaling pathways were validated in plasma samples. Alphafetoprotein (AFP), metalloproteinase (MMP)1, osteopontin (OPN), and pregnancy-specific beta-1-glycoprotein (PSG)9 were significantly increased in HCC patients. The overall performance of MMP1 and OPN in the diagnosis of HCC remained greater than that of AFP. In addition, this study represents the first report of MMP1 as a biomarker with a higher sensitivity and specificity than AFP. Thus, this study provides a valuable resource of the HCC secretome with the potential to investigate serological biomarkers. MMP1 and OPN could be used as novel biomarkers for the early detection of HCC and to improve the sensitivity of biomarkers compared with AFP.
Article PDF
Similar content being viewed by others
Avoid common mistakes on your manuscript.
References
Jemal A, Bray F, Center M M, et al. Global cancer statistics. CA Cancer J Clin, 2011, 61: 69–90
Liang X, Bi S, Yang W, et al. Epidemiological serosurvey of hepatitis B in China-declining HBV prevalence due to hepatitis B vaccination. Vaccine, 2009, 27: 6550–6557
Portolani N, Coniglio A, Ghidoni S, et al. Early and late recurrence after liver resection for hepatocellular carcinoma: prognostic and therapeutic implications. Ann Surg, 2006, 243: 229–235
Shimada K, Sakamoto Y, Esaki M, et al. Analysis of prognostic factors affecting survival after initial recurrence and treatment efficacy for recurrence in patients undergoing potentially curative hepatectomy for hepatocellular carcinoma. Ann Surg Oncol, 2007, 14: 2337–2347
Mao Y, Yang H, Xu H, et al. Golgi protein 73 (GOLPH2) is a valuable serum marker for hepatocellular carcinoma. Gut, 2010, 59: 1687–1693
States D J, Omenn G S, Blackwell T W, et al. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat Biotechnol, 2006, 24: 333–338
Meng R, Gormley M, Bhat V B, et al. Low abundance protein enrichment for discovery of candidate plasma protein biomarkers for early detection of breast cancer. J Proteomics, 2011, 75: 366–374
Makridakis M, Vlahou A. Secretome proteomics for discovery of cancer biomarkers. J Proteomics, 2010, 73: 2291–2305
Planque C, Kulasingam V, Smith C R, et al. Identification of five candidate lung cancer biomarkers by proteomics analysis of conditioned media of four lung cancer cell lines. Mol Cell Proteomics, 2009, 8: 2746–2758
Polisetty R V, Gupta M K, Nair S C, et al. Glioblastoma cell secretome: analysis of three glioblastoma cell lines reveal 148 non-redundant proteins. J Proteomics, 2011, 74: 1918–1925
Xiao T, Ying W, Li L, et al. An approach to studying lung cancer-related proteins in human blood. Mol Cell Proteomics, 2005, 4: 1480–1486
Zhang Y, Xu B, Liu Y, et al. The ovarian cancer-derived secretory/releasing proteome: a repertoire of tumor markers. Proteomics, 2012, 12: 1883–1891
Lechner J F, LaVeck M A. A serum-free method for culturing normal human bronchial epithelial cells at clonal density. J Tissue Cult Methods, 1985, 9: 43–48
Shen Z, Li P, Ni R J, et al. Label-free quantitative proteomics analysis of etiolated maize seedling leaves during greening. Mol Cell Proteomics, 2009, 8: 2443–2460
Sun H, Fang H, Chen T, et al. GOFFA: gene ontology for functional analysis—a FDA gene ontology tool for analysis of genomic and proteomic data. BMC Bioinformatics, 2006, 7(Suppl 2): S23
Wu S, Wan P, Li J, et al. Multi-modality of pI distribution in whole proteome. Proteomics, 2006, 6: 449–455
Omenn G S, States D J, Adamski M, et al. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics, 2005, 5: 3226–3245
Anderson N L, Polanski M, Pieper R, et al. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol Cell Proteomics, 2004, 3: 311–326
Chinese Human Liver Proteome Profiling Consortium. First insight into the human liver proteome from PROTEOME(SKY)-LIVER(Hu) 1.0, a publicly available database. J Proteome Res, 2010, 9: 79–94
Lin L, Amin R, Gallicano G I, et al. The STAT3 inhibitor NSC 74859 is effective in hepatocellular cancers with disrupted TGF-beta signaling. Oncogene, 2009, 28: 961–972
Pavlou M P, Diamandis E P. The cancer cell secretome: a good source for discovering biomarkers? J Proteomics, 2010, 73: 1896–1906
Sun S, Xu M Z, Poon R T, et al. Circulating Lamin B1 (LMNB1) biomarker detects early stages of liver cancer in patients. J Proteome Res, 2010, 9: 70–78
Luk J M, Lam C T, Siu A F, et al. Proteomic profiling of hepatocellular carcinoma in Chinese cohort reveals heat-shock proteins (Hsp27, Hsp70, GRP78) up-regulation and their associated prognostic values. Proteomics, 2006, 6: 1049–1057
Murata M, Matsuzaki K, Yoshida K, et al. Hepatitis B virus X protein shifts human hepatic transforming growth factor (TGF)-beta signaling from tumor suppression to oncogenesis in early chronic hepatitis B. Hepatology, 2009, 49: 1203–1217
Takafuji V, Forgues M, Unsworth E, et al. An osteopontin fragment is essential for tumor cell invasion in hepatocellular carcinoma. Oncogene, 2007, 26: 6361–6371
Shang S, Plymoth A, Ge S, et al. Identification of osteopontin as a novel marker for early hepatocellular carcinoma. Hepatology, 2012, 55: 483–490
Sun J, Xu H M, Zhou H J, et al. The prognostic significance of preoperative plasma levels of osteopontin in patients with TNM stage-I of hepatocellular carcinoma. J Cancer Res Clin Oncol, 2010, 136: 1–7
Salahshor S, Goncalves J, Chetty R, et al. Differential gene expression profile reveals deregulation of pregnancy specific beta1 glycoprotein 9 early during colorectal carcinogenesis. BMC Cancer, 2005, 5: 66
Lisboa F A, Warren J, Sulkowski G, et al. Pregnancy-specific glycoprotein 1 induces endothelial tubulogenesis through interaction with cell surface proteoglycans. J Biol Chem, 2011, 286: 7577–7586
Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell, 2010, 141: 52–67
Asselah T, Bieche I, Laurendeau I, et al. Liver gene expression signature of mild fibrosis in patients with chronic hepatitis C. Gastroenterology, 2005, 129: 2064–2075
Liu D, Guo H, Li Y, et al. Association between polymorphisms in the promoter regions of matrix metalloproteinases (MMPs) and risk of cancer metastasis: a meta-analysis. PLoS ONE, 2012, 7: e31251
Decock J, Hendrickx W, Vanleeuw U, et al. Plasma MMP1 and MMP8 expression in breast cancer: protective role of MMP8 against lymph node metastasis. BMC Cancer, 2008, 8: 77
Li M, Xiao T, Zhang Y, et al. Prognostic significance of matrix metalloproteinase-1 levels in peripheral plasma and tumour tissues of lung cancer patients. Lung Cancer, 2010, 69: 341–347
Author information
Authors and Affiliations
Corresponding authors
Additional information
This article is published with open access at Springerlink.com
Electronic supplementary material
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Yang, L., Rong, W., Xiao, T. et al. Secretory/releasing proteome-based identification of plasma biomarkers in HBV-associated hepatocellular carcinoma. Sci. China Life Sci. 56, 638–646 (2013). https://doi.org/10.1007/s11427-013-4497-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-013-4497-x