Abstract
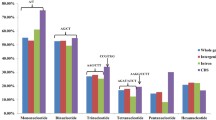
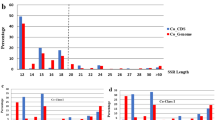
We mapped and analyzed the microsatellites throughout 284295605 base pairs of the unambiguously assembled sequence scaffolds along 19 chromosomes of the haploid poplar genome. Totally, we found 150985 SSRs with repeat unit lengths between 2 and 5 bp. The established microsatellite physical map demonstrated that SSRs were distributed relatively evenly across the genome of Populus. On average, These SSRs occurred every 1883 bp within the poplar genome and the SSR densities in intergenic regions, introns, exons and UTRs were 85.4%, 10.7%, 2.7% and 1.2%, respectively. We took di-, tri-, tetra-and pentamers as the four classes of repeat units and found that the density of each class of SSRs decreased with the repeat unit lengths except for the tetranucleotide repeats. It was noteworthy that the length diversification of microsatellite sequences was negatively correlated with their repeat unit length and the SSRs with shorter repeat units gained repeats faster than the SSRs with longer repeat units. We also found that the GC content of poplar sequence significantly correlated with densities of SSRs with uneven repeat unit lengths (tri-and penta-), but had no significant correlation with densities of SSRs with even repeat unit lengths (di-and tetra-). In poplar genome, there were evidences that the occurrence of different microsatellites was under selection and the GC content in SSR sequences was found to significantly relate to the functional importance of microsatellites.
Similar content being viewed by others
References
Wullschleger S D, Jansson S, Taylor G. Genomics and forest biology: Populus emerges as the perennial favortie. Plant Cell, 2002, 14: 2651–2655
Bradshaw H D, Ceulemans R, Davis J, et al. R. Emerging model systems in plant biology: Poplar (Populus) as a model torest tree. J Plant Growth Regul, 2000, 19: 306–313
Tuskan G A, DiFazio S P, Teichmann T. Poplar genomics is getting popular: The impact of the poplar genome project on tree research. Plant Biol, 2004, 6: 2–4
Brunner A M, Busov V B, Strauss S H. Poplar genome sequence: Functional genomics in an ecologically dominant plant species. Trends Plant Sci, 2004, 9: 49–56
Tuskan G A, DiFazio S P, Hellsten U, et al. The genome of black cottonwood, Populus Trichocarpa (Torr.& Gray Ex Brayshaw). Science, 2006, 313: 1596–1604
Schlotterer C. Genealogical inference of closely related species based on microsatellites. Genet Res Camb, 2001, 78: 209–212
Powell W, Machray G C, Provan J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci, 1996, 1: 215–222
Jewell E, Robinson A, Savage D, et al. SSR primer and SSR taxonomy tree: Biome SSR discovery. Nucl Acids Res, 2006, 34: W656–W659
Tuskan G A, Gunter L E, Yang Z M, et al. Characterization of microsatellites revealed by genomci sequencing of Populus Trichocarpa. Can For Res, 2004, 34: 85–93
Wyman J, Bruneau A, Tremblay M F. Microsatellite analysis of gentic diversity in four populations of Populus tremuloides in Quebec. Can J Bot, 2003, 81: 367
Yin T M, Difazio S P, Gunter L E, et al. Large-scale heterospecific segregation distortion in Populus revealed by a Dense Genetic Map. Theor Appl Genet, 2004, 109: 451–463
Tautz D, Trick M, Dover G A. Cryptic simplicity in DNA is a major source of genetic variation. Nature, 1986, 322: 652–656
Kashi Y, King D, Soller M. Simple sequence repeats as a source of quantitative genetic variation. Trends Genet, 1997, 13: 74–78
Li Y C, Korol A B, Beiles A, et al. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Mol Ecol, 2002, 11: 2453–2465
Dieringer D, Schlotterer C. Two distinct modes of microsatellite mutation processes: Evidence from the complete genomic sequences of nine species. Genome Res, 2003, 13: 2242–2251
Voorrips R E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J Heredity, 2002, 93: 77–78
Smith E C. A study of cytology and speciation in the Genus Populus L. J Arnold Arb, 1943, 24: 275 305
Toth G, Gaspari Z, Jurka, J. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Res, 2000, 10: 967–981
Hancock J M. Simple sequences in a ‘minimal’ genome. Nat Genet, 1996, 14: 14–15
Brinkmann B, Klintschar M, Neuhuber F, et al. Mutation rate in hu-man microsatellites: Influence of the structure and length of the tandem repeat. Am J Hum Genet, 1998, 62: 1408–1415
Brohede J, Primmer C R, Moller A, et al. Heterogeneity in the rate and pattern of germline mutation at individual microsatellite loci. Nucl Acids Res, 2002, 30: 1997–2003
Shinde D, Lai Y, Sun F, et al. Taq DNA polymerase slippage mutation rates measured by PCR and quasi-likelihood analysis: (CA/GT)n and (A/T)n microsatellites. Nucleic Acids Res, 2003, 31: 974–980
Bachtrog D, Agis M, Imhof M, et al. Microsatellite variability differs between dinucleotide repeat motifs—Evidence from Drosophila melanogaster. Mol Biol Evol, 2000, 17: 1277–1285
Hancock J M. The contribution of slippage-like processes to genome evolution. J Mol Evol, 1995, 41: 1038–1047
Reddy P S, Housman D E. The complex pathology of trinucleotide repeats. Curr Opin Cell Biol, 1997, 9: 364–372
Lothe R A. Microsatellite instability in human solid tumors. Mol Med Today, 1997, 3: 61–68
The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 2000, 408: 796–815
Goff S A, Ricke D, Lan T H, et al. A draft sequence of the rice genome (Oryza Sativa L. Ssp. Japonica). Science, 2002, 296: 92–100
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by Program for New Century Excellent Talents in University (Grant No. NCET-04-0516), Fok Ying Tung Education Foundation, and the National Natural Science Foundation of China (Grant No. 30200224)
Rights and permissions
About this article
Cite this article
Li, S., Yin, T. Map and analysis of microsatellites in the genome of Populus: The first sequenced perennial plant. SCI CHINA SER C 50, 690–699 (2007). https://doi.org/10.1007/s11427-007-0073-6
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s11427-007-0073-6