Abstract
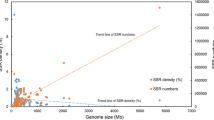
Simple sequences present in long (>30 kb) sequences representative of the single-copy genome of five species (Homo sapiens, Caenorhabditis elegans Saccharomyces cerevisiae, E. coli, and Mycobacterium leprae) have been analyzed. A close relationship was observed between genome size and the overall level of sequence repetition. This suggested that the incorporation of simple sequences had accompanied increases of genome size during evolution. Densities of simple sequence motifs were higher in noncoding regions than in coding regions in eukaryotes but not in eubacteria. All five genomes showed very biased frequency distributions of simple sequence motifs in all species, particularly in eukaryotes where AAA and TTT predominated. Interspecific comparisons showed that noncoding sequences in eukaryotes showed highly significantly similar frequency distributions of simple sequence motifs but this was not true of coding sequences. ANOVA of the frequency distributions of simple sequence motifs indicated strong contributions from motif base composition and repeat unit length, but much of the variation remained unexplained by these parameters. The sequence composition of simple sequences therefore appears to reflect both underlying sequence biases in slippage-like processes and the action of selection. Frequency distributions of simple sequence motifs in coding sequences correlated weakly or not at all with those in noncoding sequences. Selection on coding sequences to eliminate undesirable sequences may therefore have been strong, particularly in the human lineage.
Similar content being viewed by others
References
Bebenek K, Kunkel TA (1990) Frameshift errors initiated by nucleotide misincorporation. Proc Natl Acad Sci USA 87:4946–4950
Behn-Krappa A, Doerfler W (1994) Enzymatic amplification of synthetic oligodeoxyribonucleotides—implications for triplet repeat expansions in the human genome. Hum Mutat 3:19–24.
Benson DA, Boguski M, Lipman DJ, Ostell J (1994) GenBank. Nucleic Acids Res 22:3441–3444
Cavalier-Smith T (1978) Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth rate, and the solution of the DNA C-value paradox. J Cell Sci 34:247–278
Cavalier-Smith T (1985) Eukaryotic gene numbers, non-coding DNA and genome size. In: Cavalier-Smith T (ed) The evolution of genome size. John Wiley, New York, pp 69–103
Daniels DL, Plunkett III G, Burland V, Blattner FR (1992) Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science 257:771–778
Dujon B, 107 others (1994) Complete DNA sequence of yeast chromosome XI. Nature 369:371–378
Eiglmeier K, Honore N, Woods SA (1993) Use of an ordered cosmid library to deduce the genomic organization of Mycobacterium leprae. Mol Microbiol 7:197–206
Genetics Computer Group (1994) Program manual for the Wisconsin package, version 8. Genetics Computer Group, Madison, WI
Hancock JM (1993) Evolution of sequence repetition and gene duplications in the TATA-binding protein TBP (TFIID). Nucleic Acids Res 21:2823–2830
Hancock JM (1995) The contribution of DNA slippage to eukaryotic nuclear 18S rRNA evolution. J Mol Evol 40:629–639
Hancock JM, Armstrong JS (1994) SIMPLE34: an improved and enhanced implementation for VAX and Sun computers of the SIMPLE algorithm for analysis of clustered repetitive motifs in nucleotide sequences. Comput Appl Biosci 10:67–70
Hancock JM, Dover GA (1988) Molecular coevolution among cryptically simple expansion segments of eukaryotic 26S/28S rRNAs. Mol Biol Evol 5:377–391
Hancock JM, Dover GA (1990) Compensatory slippage in the evolution of ribosomal RNA genes. Nucleic Acids Res 18:5949–5954
Hoelzel AR, Hancock JM, Dover GA (1991) Evolution of the cetacean mitochondrial D-loop region. Mol Biol Evol 8:475–493
Hoelzel AR, Hancock JM, Dover GA (1993) Generation of VNTRs and heteroplasmy by sequence turnover in the mitochondrial control region of two elephant seal species. J Mol Evol 37:190–197
Jain HK (1980) Incidental DNA. Nature 288:647–648.
Jeffreys AJ, Tamaki K, MacLeod A, Monckton DG, Neil DL, Armour JAL (1994) Complex gene conversion events in germline mutation at human microsatellites. Nature Genet 6:136–145
Jurka J, Walichiewicz J, Miloslavljevic A (1992) Prototypic sequences for human repetitive DNA. J Mol Evol 35:286–291
Jurka J, Kaplan DJ, Duncan CH, Walichiewicz J, Miloslavljevic J, Miloslavljevic A, Murali G, Solus JF (1993) Identification and characterization of new human medium reiteration frequency repeats. Nucleic Acids Res 21:1273–1279
Koop BF, Hood L (1994) Striking sequence similarity over almost 100 kilobases of human and mouse T-cell receptor DNA. Nature Genet 7:48–53
Levinson G, Gutman GA (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol 4:203–221
Li T, Nicolaou C (1994) Chemical self-replication of palindromic duplex DNA. Nature 369:218–221
Lichtenauer-Kaligis EGR, Thijssen J, Den Dulk H, Van De Putte P, Tasseron-De Jong JG, Giphart-Gassler M (1993) Genome wide spontaneous mutation in human cells determined by the spectrum of mutations in hprt cDNA genes. Mutagenesis 8:207–220
Nomenclature Committee of the International Union of Biochemistry (NC-IUB) (1985) Nomenclature for incompletely specified bases in nucleic acid sequences. Eur J Biochem 150:1–5
Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448
Richards RI, Sutherland GR (1994) Simple repeat DNA is not replicated simply. Nature Genet 6:114–116
Rubinsztein DC, Amos W, Leggo J, Goodbum S, Ramesar RS, Old J, Bontrop R, McMahon R, Barton DE, Ferguson-Smith MA (1994) Mutational bias provides a model for the evolution of Huntington's disease and predicts a general increase in disease prevalence. Nature Genet 7:525–530
Rysavy FR, Bishop MJ, Gibbs GP, Williams GW (1992) The UK Human Genome Mapping Project online computing service. Comput Appl Biosci 8:149–154
Schlötterer C, Tautz D (1992) Slippage synthesis of simple sequence DNA. Nucleic Acids Res 20:211–215
Sharp PM, Lloyd AT (1993) Regional base composition variation along yeast chromosome III: evolution of chromosome primary structure. Nucleic Acids Res 21:179–183
Sievers D, von Kiedrowski G (1994) Self-replication of complementary nucleotide-based oligomers. Nature 369:221–224
Strand M, Prolla TA, Liskay RM, Petes TD (1993) Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365:274–276
Streisinger G, Okada Y, Emrich J, Newton A, Tsugita E, Terzhaghi E, Inouye M (1966) Frameshift mutations and the genetic code. Cold Spring Harbor Symp Quant Biol 31:77–84
Tautz D, Renz M (1984) Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res 12:4127–4138
Tautz D, Trick M, Dover GA (1986) Cryptic simplicity in DNA is a major source of genetic variation. Nature 322:652–656
Treier M, Pfeifle C, Tautz D (1989) Comparison of the gap segmentation gene hunchback between Drosophila melanogaster and Drosophila virilis reveals novel modes of evolutionary change. EMBO J 8:1517–1525
Valle G (1993) TA-repeat microsatellites are closely associated with ARS consensus sequences in yeast chromosome III. Yeast 9:753–759
Wilson R, 54 others (1994) 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature 368:32–38
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Hancock, J.M. The contribution of slippage-like processes to genome evolution. J Mol Evol 41, 1038–1047 (1995). https://doi.org/10.1007/BF00173185
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00173185