Abstract
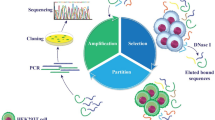
Liver cancer, as the second cause of cancer death all around the world, resulted in a series of chronic liver diseases. More than 80% of the patients cannot receive effective treatment because of their advanced disease or poor liver function. It is time to improve early clinical diagnosis and find optimal therapeutic treatments. As the tumor cells behave differently from the cell-surface molecules, it is necessary to find a highly specific probe. The aptamers, known as “chemical antibodies”, can bind to their target molecules with high affinity and high specificity. The apatmers were obtained by Cell-SELEX, which was aimed at finding the aptamers against whole living cells. In the article, after 19 selections, the ssDNA pool was cloned and sequenced. After that, six aptamers were obtained, named apt_A to apt_F. By incubating the aptamers with different cells, except apt_E, the other aptamers showed high specificity. As for apt_E, which showed high affinity to several cancer cells, was a potential probe for the common protein presented by several different cancer cells. The equilibrium dissociation constants (Kd) were evaluated by measuring the flow cytometry signal that characterized the binding ability of aptamers to the target cells at a series of concentrations ranging from 46.3(4.5) nM to 199.4(44.2) nM, which exposed the high binding affinities of these aptamers. The research in the confocal fluorescence images further confirmed the specificity of these aptamers and the fact that the aptamers were combined with the targets on the cell-surface.
Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. CA-Cancer J Clin, 2015, 65: 5–29
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. CA-Cancer J Clin, 2015, 65: 87–108
Tsukuma H, Hiyama T, Tanaka S, Nakao M, Yabuuchi T, Kitamura T, Nakanishi K, Fujimoto I, Inoue A, Yamazaki H, Kawashima T. N Engl J Med, 1993, 328: 1797–1801
Smith BD, Jorgensen C, Zibbell JE, Beckett GA. Clin Infect Disease, 2012, 55: S49–S53
Graf D, Vallböhmer D, Knoefel WT, Kröpil P, Antoch G, Sagir A, Häussinger D. Eur J Internal Med, 2014, 25: 430–437
Lencioni R, Crocetti L. Radiology, 2012, 262: 43–58
Brown S R, Martin R C G. Minerva Chir, 2012, 67: 297–308
Zhang W, Yu Y, Fang Y, Wang Y, Cui Y, Shen K, Liu T. Clin Transl Oncol, 2015, 17: 888–894
Dueland S, Guren TK, Hagness M, Glimelius B, Line PD, Pfeiffer P, Foss A, Tveit KM. Ann Surg, 2015, 261: 956–960
Rickenbacher A, DeOliveira ML, Tian Y, Jang JH, Riener MO, Graf R, Moritz W, Clavien PA. Liver Int, 2011, 31: 313–321
Wang Y, Zhang W, Qian S, Liu R, Kan Z, Wang JH. J Int Med Res, 2012, 40: 1141–1148
Zhang Y, Li Q, Wang J, Cheng F, Huang X, Cheng Y, Wang K. Cancer Lett, 2016, 377: 117–125
Shan B, Dong M, Tang H, Wang N, Zhang J, Yan C Q, Jiao X C, Zhang H L, Wang C. Oncol Lett, 2014, 8: 345–350
Liu M, Li Z, Yang J, Jiang Y, Chen Z, Ali Z, He N, Wang Z. Cell Prolif, 2016, 49: 409–420
Bunka DHJ, Stockley PG. Nat Rev Micro, 2006, 4: 588–596
Xi Z, Huang R, Deng Y, He N. J Biomed Nanotechnol, 2014, 10: 3043–3062
Romig TS, Bell C, Drolet DW. J Chromatogr B-Biomed Sci Appl, 1999, 731: 275–284
Kopylov AM. Mol Biol, 2000, 34: 940–954
Li M, Zhou X, Ding W, Guo S, Wu N. Biosens Bioelectron, 2013, 41: 889–893
Wang RE, Zhang Y, Cai J, Cai W, Gao T. Curr Med Chem, 2011, 18: 4175–4184
Yan L, Chang YN, Yin W, Tian G, Zhou L, Liu X, Xing G, Zhao L, Gu Z, Zhao Y. Biomater Sci, 2014, 2: 1412–1418
Sung T C, Chen W Y, Shah P, Chen C S. Sci Rep, 2016, 6: 20369
Lao YH, Chi CW, Friedrich SM, Peck K, Wang TH, Leong KW, Chen LC. ACS Appl Mater Interfaces, 2016, 8: 12048–12055
Zhou ZM, Feng Z, Zhou J, Fang BY, Ma ZY, Liu B, Zhao YD, Hu XB. Sensor Actuat B-Chem, 2015, 210: 158–164
Wang YX, Ye ZZ, Ying YB. Chin Sci Bull, 2013, 58: 2938–2943
Rockey WM, Huang L, Kloepping KC, Baumhover NJ, Giangrande PH, Schultz MK. Bioorg Medicinal Chem, 2011, 19: 4080–4090
Kang ST, Luo YL, Huang YF, Yeh CK. IEEE Int Ultrason Symp, 2012: 1866–1868
Medley CD, Bamrungsap S, Tan W, Smith JE. Anal Chem, 2011, 83: 727–734
Kim D, Jeong YY, Jon S. ACS Nano, 2010, 4: 3689–3696
Kong RM, Chen Z, Ye M, Zhang XB, Tan WH. Sci China Chem, 2011, 54: 1218–1226
Ma F, Jia LJ, Zhang Y, Sun B, Qi HL, Gao Q, Zhang CX. Sci China Chem, 2011, 54: 1357–1364
Xi Z, Huang R, Li Z, He N, Wang T, Su E, Deng Y. ACS Appl Mater Interfaces, 2015, 7: 11215–11223
Xi Z, Zheng B, Wang C. Nanosci Nanotechnol Lett, 2016, 8: 1061–1066
Salimi A, Khezrian S, Hallaj R, Vaziry A. Anal Biochem, 2014, 466: 89–97
Kavosi B, Salimi A, Hallaj R, Moradi F. Biosens Bioelectron, 2015, 74: 915–923
Subramanian N, Raghunathan V, Kanwar J R, Kanwar R K, Elchuri S V, Khetan V, Krishnakumar S. Mol Vis, 2012, 18: 2783–2795
Wang RW, Zhu G, Mei L, Xie Y, Ma H, Ye M, Qing FL, Tan W. J Am Chem Soc, 2014, 136: 2731–2734
Zhang Z, Ali MM, Eckert MA, Kang DK, Chen YY, Sender LS, Fruman DA, Zhao W. Biomaterials, 2013, 34: 9728–9735
Shin SW, Song WC, Kim AR, Cho SW, Kim DI, Um SH. Biomater Sci, 2014, 2: 76–83
Sun W, Gu Z. Biomater Sci, 2015, 3: 1018–1024
Huang R, Xi Z, He N. Sci China Chem, 2015, 58: 1122–1130
Berezhnoy A, Brenneman R, Bajgelman M, Seales D, Gilboa E. Mol Ther-Nucl Acids, 2012, 1: e51
Subramanian N, Kanwar JR, Athalya PK, Janakiraman N, Khetan V, Kanwar RK, Eluchuri S, Krishnakumar S. J Biomed Sci, 2015, 22: 4
Mascini M. Anal Bioanal Chem, 2008, 390: 987–988
Dua P, Kim S, Lee DK. Methods, 2011, 54: 215–225
Guo KT, Ziemer G, Paul A, Wendel HP. Int J Mol Sci, 2008, 9: 668–678
Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proc Natl Acad Sci USA, 2006, 103: 11838–11843
Zhao J, Lee SH, Huss M, Holme P. Sci Rep, 2013, 3: 1583
Acknowledgments
This work was supported by the National Science Foundation of China (61471168, 61571187, 61301043, 61527806), the China Postdoctoral Science Foundation (2016T90403) and the Economical Forest Cultivation and Utilization of 2011 Collaborative Innovation Center in Hunan Province [(2013) 448].
Author information
Authors and Affiliations
Corresponding authors
Additional information
These authors contributed equally to this work.
Rights and permissions
About this article
Cite this article
Huang, R., Chen, Z., Liu, M. et al. The aptamers generated from HepG2 cells. Sci. China Chem. 60, 786–792 (2017). https://doi.org/10.1007/s11426-016-0491-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11426-016-0491-7