Abstract
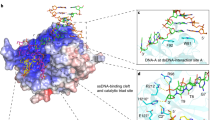
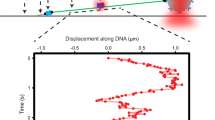
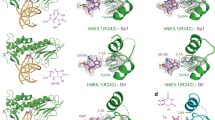
Mammalian AlkB homologue 2 (ALKBH2) is the primary housekeeping DNA demethylase, effectively repairing endogenously formed methylated lesions in double-stranded DNA. Our previous studies demonstrated that a hydrophobic β-hairpin motif of ALKBH2 could play crucial roles in base-pair stability interrogation and damaged base flipping. Using chemical cross-linking strategy, we obtained two crystal structures of human ALKBH2 mutant bound to duplex DNA. The structural analysis suggests that the β-hairpin motif is flexible in conformation and is likely to slide along the DNA duplex in local regions to search for damaged base. This study provides a new mechanistic insight into DNA damage detection by ALKBH2.
Similar content being viewed by others
References
Sedgwick B. Repairing DNA-methylation damage. Nat Rev Mol Cell Biol, 2004, 5(2): 148–157
Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem, 1988, 57: 133–157
Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: Recent advances. DNA Repair (Amst), 2007, 6(4): 429–442
Mishina Y, Duguid EM, He C. Direct reversal of DNA alkylation damage. Chem Rev, 2006, 106(2): 215–232
Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature, 2002, 419(6903): 178–182
Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, Krokan HE. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature, 2003, 421(6925): 859–863
Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci USA, 2002, 99(26): 16660–16665
Ringvoll J, Nordstrand LM, Vagbo CB, Talstad V, Reite K, Aas PA, Lauritzen KH, Liabakk NB, Bjork A, Doughty RW, Falnes PO, Krokan HE, Klungland A. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. Embo J, 2006, 25(10): 2189–2198
Gilljam KM, Feyzi E, Aas PA, Sousa MM, Muller R, Vagbo CB, Catterall TC, Liabakk NB, Slupphaug G, Drablos F, Krokan HE, Otterlei M. Identification of a novel, widespread, and functionally important PCNA-binding motif. J Cell Biol, 2009, 186(5): 645–654
Fujii T, Shimada K, Anai S, Fujimoto K, Konishi N. ALKBH2, a novel AlkB homologue, contributes to human bladder cancer progression by regulating MUC1 expression. Cancer Sci, 2013, 104(3): 321–327
Cetica V, Genitori L, Giunti L, Sanzo M, Bernini G, Massimino M, Sardi I. Pediatric brain tumors: mutations of two dioxygenases (hABH2 and hABH3) that directly repair alkylation damage. J Neurooncol, 2009, 94(2): 195–201
Johannessen TC, Prestegarden L, Grudic A, Hegi ME, Tysnes BB, Bjerkvig R. The DNA repair protein ALKBH2 mediates temozolomide resistance in human glioblastoma cells. Neuro Oncol, 2013, 15(3): 269–278
Mishina Y, Chen LX, He C. Preparation and characterization of the native iron(II)-containing DNA repair AlkB protein directly from Escherichia coli. J Am Chem Soc, 2004, 126(51): 16930–16936
Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: Implications for drug resistance. Science, 1998, 282(5394): 1669–1675
Verdine GL, Norman DP. Covalent trapping of protein-DNA complexes. Annu Rev Biochem, 2003, 72(337–366)
Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science, 2006, 311(5764): 1153–1157
Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature, 2008, 452(7190): 961–965
Yi C, Chen B, Qi B, Zhang W, Jia G, Zhang L, Li CJ, Dinner AR, Yang CG, He C. Duplex interrogation by a direct DNA repair protein in search of base damage. Nat Struct Mol Biol, 2012, 19(7): 671–676
Chen B, Liu H, Sun X, Yang CG. Mechanistic insight into the recognition of single-stranded and double-stranded DNA substrates by ABH2 and ABH3. Mol Biosyst, 2010, 6(11): 2143–2149
Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol, 1997, 276: 307–326
The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr, 1994, 50(Pt 5): 760–763
Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr, 2001, 57(Pt 10): 1373–1382
Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr, 2004, 60(Pt 12 Pt 1): 2126–2132
Yi C, Yang CG, He C. A non-heme iron-mediated chemical demethylation in DNA and RNA. Acc Chem Res, 2009, 42(4): 519–529
Monsen VT, Sundheim O, Aas PA, Westbye MP, Sousa MM, Slupphaug G, Krokan HE. Divergent ss-hairpins determine double-strand versus single-strand substrate recognition of human AlkB-homologues 2 and 3. Nucleic Acids Res, 2010, 38(19): 6447–6455
Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci U S A, 2006, 103(15): 5752–5757
Lin Y, Zhao T, Jian X, Farooqui Z, Qu X, He C, Dinner AR, Scherer NF. Using the bias from flow to elucidate single DNA repair protein sliding and interactions with DNA. Biophys J, 2009, 96(5): 1911–1917
Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry, 2010, 49(24): 4957–4967
Blainey PC, Luo GB, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically bound proteins spin while diffusing along DNA. Nat Struct Mol Biol, 2009, 16(12): U1224–U1234
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Chen, B., Gan, J. & Yang, C. The complex structures of ALKBH2 mutants cross-linked to dsDNA reveal the conformational swing of β-hairpin. Sci. China Chem. 57, 307–313 (2014). https://doi.org/10.1007/s11426-013-5029-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11426-013-5029-z