Abstract
Fungi have long been regarded as abundant sources of natural products (NPs) exhibiting significant biological activities. Decades of studies on the biosynthesis of fungal NPs revealed that most of the biosynthetic steps are catalyzed by sophisticated enzymes encoded in biosynthetic gene clusters, whereas some reactions proceed without enzymes. These non-enzymatic reactions complicate biosynthetic analysis of NPs and play important roles in diversifying the structure of the products. Therefore, knowledge on the non-enzymatic reactions is important for elucidating the biosynthetic mechanism. This review focuses on non-enzymatic reactions we recently encountered during biosynthetic studies of four types of NPs (viridicatins, Sch210972, lentopeptins, and lentofuranine).
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fungi produce a wide range of natural products (NPs) such as polyketides [1], terpenes [2], non-ribosomal peptides [3] and hybrids of them [4, 5]. These compounds sometimes exhibit significant biological activities and thus fungi have been considered as attractive sources of pesticides and pharmaceuticals. Understanding the biosynthesis of NPs allows for the discovery of novel NPs through genome mining and the creation of modified NPs via genome engineering. As a result, the biosynthetic pathways of numerous important metabolites have been extensively researched and established so far [1,2,3,4,5]. Biosynthesis of NPs is usually catalyzed by sophisticated biosynthetic enzymes but not all of the reactions require enzymes. These chemical reactions proceeding without enzymes are called non-enzymatic reactions [6, 7]. One of nature’s most famous non-enzymatic reactions is photochemical conversion of 7-dehydrocholesterol to cholecalciferol, also known as vitamin D3 (Fig. 1a) [8]. Another example can be found in the formation of artemisinin from dihydroartemisinic acid although the mechanism remains unclear (Fig. 1b) [9]. These examples show that non-enzymatic reactions are not always undesired nor unnecessary for the producer and human beings. On the other hand, non-enzymatic conversion of NPs sometimes complicates biosynthetic analysis. In this review, recent discoveries of non-enzymatic reactions in the fungal biogenesis of NPs and related enzymatic reactions are introduced and discussed.
Biosynthesis of viridicatins
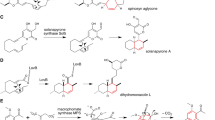
Viridicatin (1), viridicatol (2), and 4′-methoxyviridicatin (3) are fungal NPs produced by Aspergillus and Penicillium species [10,11,12]. The first report on the biosynthesis of these compounds was provided in 1967. Luckner showed that homogenized mycelia of Penicillium viridicatum could convert cyclopenin (4) and cyclopenol (5) into 1 and 2, respectively [13]. The active material in the mycelia was deduced to be an enzyme and was named “cyclopenase”, which was not identified for a long time. In 2014, Ishikawa and co-workers reported that non-heme iron dioxygenase AsqJ could produce 4 and 4′-methoxycyclopenin (6) from cyclopeptin (7) and 4′-methoxy cyclopeptin (8), respectively (Fig. 2b) [14]. In this study, spontaneous formation of 3 from 6 was observed during AsqJ reaction. To the contrary, 1 was not produced from 4 in neutral pH conditions, indicating that cyclopenase is necessary for the conversion of 4 to 1 in nature. This observation encouraged us to search for the cyclopenase gene, and we found hemocyanin-like enzyme AsqI is encoded in the gene located near asqJ. Heterologously expressed AsqI converted 4 and 6 to 1 and 3, respectively, thus AsqI was proved to be the missing cyclopenase (Fig. 2a) [15]. Crystallographic and biochemical analysis of AsqI revealed that a Zn(II) ion bound in the metal binding domain is responsible for the enzymatic reaction. Zn(II) ion acts as a Lewis acid to promote the ring opening of epoxide and induces the elimination of methylisocyanate from 4 to produce 1 (Fig. 2c). The reason why 6 spontaneously transforms to 3 but 4 does not transform to 1 without cyclopenase can be explained by the effect of the methoxy group of 6. Delocalization of a lone pair of electrons on the methoxy group leads to the spontaneous opening of the epoxide ring (Fig. 2d) [14]. In the case of 4 and 5, activation of epoxide with a strong acid (Lewis acid or Brønsted acid) can induce nonenzymatic transformation at room temperature. Bräuer and co-workers reported that 1 was also produced during the AsqJ reaction, which seems to be an accident caused by trichloroacetic acid they added after the reaction [16].
Biosynthesis of Sch210972
Sch210972 (9) is an octalin-containing fungal metabolite produced by Chaetomium globosum. The biosynthetic pathway of 9 was elucidated by Sato and co-workers via gene deletion and heterologous expression in Aspergillus nidulans in 2015 [17]. CghB (aldolase), CghC (enoyl reductase), and CghG (PKS–NRPS) produced linear-chain precursor 10, which was subsequently cyclized by CghA (Diels–Alderase) to produce 9. Both 9 and its diastereomer 11 were detected from the cghA-knockout strain, suggesting 10 was non-enzymatically transformed into endo adduct 9 and exo adduct 11 (Fig. 3b). Other octalin-containing NPs such as equisetin (12) and phomasetin (13) (Fig. 3a) were also reported to be produced as a mixture of diastereomers when the corresponding Diels–Alderase genes were knocked out [18, 19]. This non-enzymatic reaction of 10 complicated biochemical characterizations of CghA: we could not obtain substrate 10 from cghA-knockout strain and we had to distinguish enzymatic products from non-enzymatic ones for kinetic analysis. To solve the first problem, we designed and synthesized simplified substrate 14, which lacks two methyl groups and one hydroxyl group of 10 [20]. Spontaneous transformation of 14 to endo-cyclization product 15 and exo-cyclization product 16 was also observed during isolation, urging us to synthesize 14 just before use and use 14 without purification. Suzuki–Miyaura cross-coupling of alkenyl iodide 17 and boronic ester 18 was chosen as the last step of the synthetic scheme because this reaction creates carbon–carbon bonds in a highly regioselective manner and can be conducted in aqueous conditions (Fig. 3c).
After completion of the synthesis of 14, point mutants of CghA were created based on the crystal structure of 9-bound CghA, and their kinetic parameters including stereoselectivity were evaluated. In this step, the stereoselectivity of the mutants was determined at first using sufficient amounts of enzymes to prevent spontaneous reactions of 14. Small amounts of enzymes were used for kinetic analysis to keep most of 14 unreacted, resulting in nonenzymatic production of 15 and 16 during HPLC analysis. To distinguish enzymatic products from nonenzymatic ones, we paid attention to the fact that the ratio of nonenzymatically produced 15 and 16 is 50:50. This means the variance observed between the quantities of 15 and 16 in the kinetic assay is attributed to the enzymatic products. Based on these data, the amounts of enzymatically produced 15 and 16 were calculated and kinetic parameters were determined. At the end of the work, two triple-mutants (A242S/M257V/V391L, A242N/M257V/V391L) with reversed stereoselectivity were obtained (Fig. 3c) [20]. This was the first report proving and changing the stereoselectivity of octalin-forming Diels–Alderase in the world.
Biosynthesis of lentopeptins
Lentopeptin A (19) and B (20), produced by Aspergillus lentulus, possess the same planar structure but differ in stereochemistry at C-2 and C-9 (Fig. 4a) [21]. Although the structure of lentopeptins resembles that of ergotamine (21), 21 is produced as a single isomer in Claviceps purpurea (Fig. 4b) [22]. To prove what makes the difference between lentopeptins and 21, the biosynthetic mechanism of lentopeptins was investigated. Knockout experiments revealed that the biosynthetic gene cluster (BGC) for lentopeptins is composed of only three genes: lenA (NRPS), lenB (phenylalanine-ammonia lyase) and lenC (P450). The biosynthesis of lentopeptins begins with the production of cinnamic acid by LenB and LenA produces mono-cyclic intermediate lentopeptin C (22) from cinnamic acid, L-alanine, and L-valine. Similar intermediate 23 is also produced in the biosynthesis of 21. However, the construction of the characteristic N-acyl diketopiperazine moiety differs between 22 and 23. Formation of 22 requires catalysis by terminal condensation (CT) domain of LenA at the cyclization step but 23 does not (Fig. 4) [23]. The proline residue in the linear precursor of 23 anchors the C-terminal thioester in proximity to the amide nitrogen, promoting spontaneous cyclization. In the last step of biosynthesis, 23 is converted only to 21 by non-heme iron dioxygenase EasH, while 22 is converted to both 19 and 20 by P450 LenC. The reaction catalyzed by LenC was examined in detail to reveal that the source of the oxygen atom incorporated during the transformation is different between 19 and 20. In the formation of 19, molecular oxygen is activated and added to the α-position of Ala residue in 22 to produce linear precursor 24, which spontaneously cyclizes to form 19 in aqueous conditions. On the other hand, one of the oxygen atoms of 20 was derived from water, suggesting that 22 was dehydrogenated and hydrated to form 20.
Biosynthesis of fumimycin and lentofuranine
Fumimycin (25), a fungal metabolite containing an unusual carbon–carbon bond between the α-carbon of alanine and an aromatic ring, was isolated from Aspergillus fumisynnematus in 2007 [24]. This unique structure attracted organic chemists around the world and asymmetric total synthesis of 25 was accomplished in 2010 [25]. Surprisingly, the optical rotation of natural 25 was much smaller than that of optical pure 25, suggesting the biosynthesis of 25 involves a spontaneous racemization step. In 2023, our group isolated 25 and structurally-related compound lentofuranine (26) from A. lentulus and Aspergillus novofumigatus [26]. The stereochemistry of 26 was determined using Marfey’s method [27], revealing that 26 was also a racemic compound. These observations encouraged us to unveil the atypical biogenesis of 25 and 26. However, we found no single BGC corresponding to synthesizing all the structures of 25 and 26 in the genome of A. lentulus and A. novofumigatus, indicating they are collaboratively synthesized by separated BGCs. Since 25 and 26 had the same aromatic moiety, PKS genes shared between A. lentulus and A. novofumigatus were knocked out to discover their BGC. One of the candidates was AlterA, which is similar to a gene corresponding to producing terrein (27) in Aspergillus terreus [28,29,30]. Deletion of AlterA in A. lentulus abolished the production of 25, 26, and 27, revealing these three compounds share the same biosynthetic origin (Fig. 5a). Knockout analysis of the other genes in the BGC unveiled that only three genes named AlterA (PKS), AlterB (PKS), and AlterC (flavin-dependent monooxygenase) are indispensable for producing 25 and 26. Judged from the function of these three, other genes located outside the BGC seemed to be necessary for the production of 25 and 26. A gene named AlsidE was raised as a candidate because its ortholog sidE was reported to produce fumarylalanine (28), a compound resembling the peptidic portion of 25 and 26 [31]. As we expected, the AlsidE-deletion strain of A. lentulus could not produce 25 and 26.
a Overview of the biosynthesis of fumimycin (25), lentofuranine (26) and terrein (27). b Nonenzymatic formation of fumimycin analog 30 using compound 29. c Unusual tautomerization of 31 to tauto-31. d Putative mechanisms of formation of 25 and 26 via nucleophilic attack from C4 (PATH C4) and C7 (PATH C7), respectively
The remaining question was how AlSidE, an NRPS with A-T-C-A-T-CT topology, produces these compounds. This was answered by in vitro analysis of AlSidE using compound 29 as an alternative to the quinone product of Alter cluster. AlSidE produced fumimycin analog 30 in addition to 28 when 29 was included in the reaction mixture. The formation of 30 was also observed in the combination of the ultrafiltrate of the AlSidE reaction mixture and 29 but not in the combination of 28 and 29 (Fig. 5b), indicating that AlSidE produced a reactive material other than 28. We hypothesized that the reactive substance was fumarylazlactone (31) due to three reasons mentioned below. First, AlSidE has a CT domain which is usually involved in the cyclization step of NRPS. Second, an azlactone is easily hydrolyzed to form a corresponding carboxylic acid in general. Third, an azlactone is known to racemize rapidly and 28 produced by AlSidE was racemic. To prove our hypothesis, we synthesized 31 as an authentic standard and found that AlSidE exactly produced 31. In addition, 31 could be detected from the wild-type strain of A. lentulus and not from the AlsidE-deletion strain. These results clearly indicated that AlsidE is responsible for the production of 31. To our knowledge, this was the first report of azlactone-synthesizing NRPS [26]. Since azlactones are highly reactive compounds, this naturally occurring azlactone had been overlooked for a long time. Detailed analysis of the reactivity of azlactone 31 revealed that 31 could spontaneously react with 29 to form 30 (Fig. 5b). Furthermore, 31 was found to tautomerize to an oxazolone form tauto-31 (Fig. 5c), indicating both C-4 and C-7 are nucleophilic. Based on these chemical properties of 31, we proposed the mechanisms of formation of 25 and 26 as shown in Fig. 5d. Interestingly, 31 was also produced by other Aspergillus and Penicillium fungi lacking an AlterA ortholog necessary for producing 25 and 26, suggesting that 31 itself would play some roles in the lifecycle of the producer as discussed in the discovery of natural oxazolones by Rond et al. [32].
Conclusion
This mini review has highlighted four types of NP biosynthesis accompanying non-enzymatic reactions. In the case of viridicatins and Sch210972, non-enzymatic reactions caused problems in the analysis of enzymatic reactions. However, these problems could be solved by changing reaction conditions to avoid non-enzymatic ones. Hence understanding what drives spontaneous reactions and how they can be prevented are important in biosynthetic study. On the other hand, non-enzymatic reactions are essential in the biosynthesis of lentopeptins and lentofuranine. In these cases, analysis of the reactivity of enzymatically produced intermediates was the key to uncovering the true mechanism of biosynthesis. Due to the large number of NP biosyntheses remaining to be elucidated, there could be numerous undiscovered non-enzymatic reactions in nature. It is important for researchers to carefully assess what is going on during the biosynthesis.
References
Cox RJ, Skellam E, Williams K (2018) Biosynthesis of fungal polyketides. Physiology and genetics. Springer, Cham, pp 385–412
Quin MB, Flynn CM, Schmidt-Dannert C (2014) Traversing the fungal terpenome. Nat Prod Rep 31:1449–1473
Süssmuth RD, Mainz A (2017) Nonribosomal peptide synthesis—principles and prospects. Angew Chem Int Ed 56:3770–3821
Matsuda Y, Abe I (2016) Biosynthesis of fungal meroterpenoids. Nat Prod Rep 33:26–53
Kishimoto S, Hirayama Y, Watanabe K (2018) Polyketide synthase–nonribosomal peptide synthetase hybrid enzymes of fungi. Physiology and genetics. Springer, Cham, pp 367–383
Keller MA, Piedrafita G, Ralser M (2015) The widespread role of non-enzymatic reactions in cellular metabolism. Curr Opin Biotechnol 34:153–161
Bouthillette LM, Aniebok V, Colosimo DA, Brumley D, Macmillan JB (2022) Nonenzymatic reactions in natural product formation. Chem Rev 122:14815–14841
Wacker M, Holick MF (2013) Sunlight and vitamin D: a global perspective for health. Dermato-endocrinology 5:51–108
Sy LK, Brown GD (2002) The mechanism of the spontaneous autoxidation of dihydroartemisinic acid. Tetrahedron 58:897–908
Cunningham KG, Freeman GG (1953) The isolation and some chemical properties of viridicatin, a metabolic product of Penicillium viridicatum Westling. Biochem J 53:328–332
Birkinshaw JH, Luckner M, Mohammed YS, Mothes K, Stickings CE (1963) Studies in the biochemistry of micro-organisms. 114. Viridicatol and cyclopenol, metabolites of Penicillium viridicatum Westling and Penicillium cyclopium Westling. Biochem J 89:196–202
He J, Lion U, Sattler I, Gollmick FA, Grabley S, Cai J, Meiners M, Schünke H, Schaumann K, Dechert U, Krohn M (2005) Diastereomeric quinolinone alkaloids from the marine-derived fungus Penicillium janczewskii. J Nat Prod 68:1397–1399
Luckner M (1967) Zur Bildung von Chinolinalkaloiden in Pflanzen: 2. Die fermentative Umwandlung der Penicillium-Alkaloide Cyclopenin und Cyclopenol in Viridicatin und Viridicatol. Eur J Biochem 2:74–78
Ishikawa N, Tanaka H, Koyama F, Noguchi H, Wang CC, Hotta K, Watanabe K (2014) Non-heme dioxygenase catalyzes atypical oxidations of 6, 7-bicyclic systems to form the 6,6-quinolone core of viridicatin-type fungal alkaloids. Angew Chem Int Ed 53:12880–12884
Kishimoto S, Hara K, Hashimoto H, Hirayama Y, Champagne PA, Houk KN, Tang Y, Watanabe K (2018) Enzymatic one-step ring contraction for quinolone biosynthesis. Nat Commun 9:2826
Bräuer A, Beck P, Hintermann L, Groll M (2016) Structure of the dioxygenase AsqJ: mechanistic insights into a one-pot multistep quinolone antibiotic biosynthesis. Angew Chem Int Ed 55:422–426
Sato M, Yagishita F, Mino T, Uchiyama N, Patel A, Chooi YH, Goda Y, Xu W, Noguchi H, Yamamoto T, Hotta K, Houk KN, Tang Y, Watanabe K (2015) Involvement of lipocalin-like CghA in decalin-forming stereoselective intramolecular [4 + 2] cycloaddition. ChemBioChem 16:2294–2298
Kato N, Nogawa T, Hirota H, Jang JH, Takahashi S, Ahn JS, Osada H (2015) A new enzyme involved in the control of the stereochemistry in the decalin formation during equisetin biosynthesis. Biochem Biophys Res Commun 460:210–215
Kato N, Nogawa T, Takita R, Kinugasa K, Kanai M, Uchiyama M, Osada H, Takahashi S (2018) Control of the stereochemical course of [4 + 2] cycloaddition during trans-decalin formation by Fsa2-Family enzymes. Angew Chem Int Ed 57:9754–9758
Sato M, Kishimoto S, Yokoyama M, Jamieson CS, Narita K, Maeda N, Hara K, Hashimoto H, Tsunematsu Y, Houk KN, Watanabe K (2021) Catalytic mechanism and endo-to-exo selectivity reversion of an octalin-forming natural Diels–Alderase. Nat Catal 4:223–232
Kishimoto S, Matsubara Y, Watanabe K (2022) Alkaloid biosynthetic enzyme generates diastereomeric pair via two distinct mechanisms. J Am Chem Soc 144:5485–5493
Wallwey C, Li S-M (2011) Ergot alkaloids: structure diversity, biosynthetic gene clusters and functional proof of biosynthetic genes. Nat Prod Rep 28:496–510
Havemann J, Vogel D, Loll B, Keller U (2014) Cyclolization of d-lysergic acid alkaloid peptides. Chem Biol 21:146–155
Kwon Y-J, Sohn M-J, Zheng C-J, Kim W-G (2007) Fumimycin: a peptide deformylase inhibitor with an unusual skeleton produced by Aspergillus fumisynnematus. Org Lett 9:2449–2451
Gross PJ, Furche F, Nieger M, Bräse S (2010) Asymmetric total synthesis of (+)-fumimycin via 1,2-addition to ketimines. Chem Commun 46:9215–9217
Kishimoto S, Minami A, Aoki Y, Matsubara Y, Watanabe S, Watanabe K (2023) A reactive azlactone intermediate drives fungal secondary metabolite cross-pathway generation. J Am Chem Soc 145:3221–3228
Fujii K, Ikai Y, Oka H, Suzuki M, Harada K-I (1997) A nonempirical method using LC/MS for determination of the absolute configuration of constituent amino acids in a peptide: combination of Marfey’s method with mass spectrometry and its practical application. Anal Chem 69:5146–5151
Zaehle C, Gressler M, Shelest E, Geib E, Hertweck C, Brock M (2014) Terrein biosynthesis in Aspergillus terreus and its impact on phytotoxicity. Chem Biol 21:719–731
Shu X, Wei G, Qiao Y, Zhang K, Zhang J, Ai G, Tang M-C, Zhang Y, Gao S-S (2021) TerC is a multifunctional and promiscuous flavoprotein monooxygenase that catalyzes bimodal oxidative transformations. Org Lett 23:8947–8951
Kahlert L, Bernardi D, Hauser M, Lóca LP, Berlinck RGS, Skellam EJ, Cox RJ (2021) Early oxidative transformations during the biosynthesis of terrein and related natural products. Chem Eur J 27:11895–11903
Steinchen W, Lackner G, Yasmin S, Schrettl M, Dahse H-M, Haas H, Hoffmeister D (2013) Bimodular peptide synthetase SidE produces fumarylalanine in the human pathogen Aspergillus fumigatus. Appl Environ Microbiol 79:6670–6676
de Rond T, Asay JE, Moore BS (2021) Co-occurrence of enzyme domains guides the discovery of an oxazolone synthetase. Nat Chem Biol 17:794–799
Acknowledgements
The author acknowledges Prof. Kenji Watanabe (University of Shizuoka) and all the co-workers for support on this work.
Funding
This work was financially supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (JSPS KAKENHI Grant Number 19K15757 and 21K14795).
Author information
Authors and Affiliations
Contributions
Not applicable.
Corresponding author
Ethics declarations
Conflict of interest
The author declares no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kishimoto, S. Non-enzymatic reactions in biogenesis of fungal natural products. J Nat Med 78, 467–473 (2024). https://doi.org/10.1007/s11418-024-01797-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11418-024-01797-z