Abstract
The ongoing SARS-CoV-2 evolution process has generated several variants due to its continuous mutations, making pandemics more critical. The present study illustrates SARS-CoV-2 evolution and its emerging mutations in five directions. First, the significant mutations in the genome and S-glycoprotein were analyzed in different variants. Three linear models were developed with the regression line to depict the mutational load for S-glycoprotein, total genome excluding S-glycoprotein, and whole genome. Second, the continent-wide evolution of SARS-CoV-2 and its variants with their clades and divergence were evaluated. It showed the region-wise evolution of the SARS-CoV-2 variants and their clustering event. The major clades for each variant were identified. One example is clade 21K, a major clade of the Omicron variant. Third, lineage dynamics and comparison between SARS-CoV-2 lineages across different countries are also illustrated, demonstrating dominant variants in various countries over time. Fourth, gene-wise mutation patterns and genetic variability of SARS-CoV-2 variants across various countries are illustrated. High mutation patterns were found in the ORF10, ORF6, S, and low mutation pattern E genes. Finally, emerging AA point mutations (T478K, L452R, N501Y, S477N, E484A, Q498R, and Y505H), their frequencies, and country-wise occurrence were identified, and the highest event of two mutations (T478K and L452R) was observed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ongoing mutations and the rise of different variants of SARS-CoV-2, created by nature, concern public health [1]. The new variants have been generated over time from the Wuhan strain, and the variants have created a new surge in the country of origin and then transmitted to other countries [2]. The new variants, especially variants of concern (VOCs), lead to the pandemic period becoming more critical because of their transmission properties. In early 2020, researchers noted the first single amino acid (AA) change in the S-glycoprotein at position 614 (D614). The change (614G) was noted in a small number of sample sequences. It was noted that the variant is significant within a few weeks [3]. The mutation is responsible for the structural alteration of the furin-like domain region in the S-glycoprotein. Due to the conformational changes, the conformational plasticity (S1/S2 furin-like site) is also altered. Therefore, the volume of the cavity of this domain of S-glycoprotein is enlarged. At the same time, it also helps to increase the volume surrounding the cleavage site in the domain [4]. It further helps the virus S-glycoprotein interact with the host ACE2 receptor, assisting the virus to infect more hosts and spread within the host. Therefore, this mutation was able to describe an increase in infectivity and reinfectivity [5, 6]. Subsequently, several variants have been recorded with a number of mutations over time [7,8,9,10]. The continuous evolution of SARS-CoV-2 and the emergence of new variants are substantial concerns for every country. The SARS-CoV-2 evolutionary process using their phylogenetic profile and the creation of variants has been studied by several research groups [11,12,13,14,15,16]. At the same time, several researchers have studied evolutionary dynamics using the different proteins of SARS-CoV-2 [17]. Although several variants have arisen, some have higher transmission capacity, infection, reinfection, and virulent properties. WHO, CDC, and eCDC described the variants as variants of interest (VOIs) or VOC concerning their associated properties.
It is noteworthy that RNA viruses have higher mutation rates than DNA viruses and a million times higher than their hosts [18]. SARS-CoV-2 is an RNA virus that acquires mutations like other RNA viruses [19, 20]. Researchers are working on estimating the virus’ evolutionary rate and mutation burden, which is approximately 1 × 10−3 substitutions per base per year [19]. De Maio et al. (2021) observed that two mutation rates (C to U and G to U mutation rate) are extremely high in SARS-CoV-2, with the calculated rate being approximately 97% [21].
The evolution of the virus occurs because of mutations and natural selection. Different mechanisms, such as systematic viral replication, host immune escape, and transmission, help to favor the natural selection of traits [22, 23]. Several mutations occur during evolution, creating significant genetic variations [24], leading to the rise of new viral variants. In SARS-CoV-2, the most studied mutations are point mutations. Kosuge et al. (2020) studied point mutations and found the prevalence of C to U point mutations in SARS-CoV-2 [25]. Miller et al. (2021) studied point mutations in the N gene. Two point mutations were reported; the first was observed in five patients (C to T mutation) at position 29,197, and the second was noted in eight patients (C to T mutation) at position 29,200 [26]. Other than point mutations, deletions and insertion in SARS-CoV-2 have also been studied by researchers [27,28,29]. In addition to these events, frame shifting RNA events are also associated with structural alterations [30]. However, it is urgently necessary to understand the evolution pattern and mutation profile of SARS-CoV-2 variants region-wise. Such studies will provide a better understanding of the clinical and epidemiological parameters of the pandemic, such as evolving transmission events of variants, and reveal indicators of constantly evolving immunity evasion phenomena and neutralizing antibody (nAb) escape events of the variants.
In this article, five major points are explained: first, overall significant mutations in the genome and S-glycoprotein in different variants and their origin; second, continent-wide evolution of SARS-CoV-2 and its variants with their clades and divergence; third, lineage dynamics and comparison between SARS-CoV-2 lineages across different countries; fourth, gene-wise mutation patterns and genetic variability of SARS-CoV-2 variants across various geographic locations; and finally, significant AA point mutations in SARS-CoV-2 variants, their frequencies, and country-wise occurrence.
Materials and methods
We have collected the data to illustrate the evolution pattern and the mutations profile of SARS-CoV-2 variants. We employed several bioinformatics servers, tools, and statistical software for our extensive analysis and model development.
Collection of data to illustrate evolution pattern and the mutations profile of SARS-CoV-2 variants
We have collected the different data for evolution patterns and the mutations profile of SARS-CoV-2 variants from several databases and scientific or medical search engines such as PubMed [31, 32] and Web of Science [33] and Google Scholar [34] on January 2022. Different information on variants was obtained from the eCDC [35], CDC, USA [36], and WHO [37]. Simultaneously, we performed different keyword searches and found several studies on SARS-CoV-variants. The keyword search was performed using different keywords such as “variants of SARS-CoV-2,” “VOI,” “VOC,” “mutation in SARS-CoV-2,” “mutation in SARS-CoV-2 genome,” and “mutation in SARS-CoV-2 Spike,” etc. We also searched using specific keywords like “mutation in Alpha variant,” “mutation in Delta variant,” and “mutation in Omicron variant,” etc. However, we did not find any studies investigating the evolution pattern and mutation profile of SARS-CoV-2 variants in various regions.
SARS-CoV-2 variants from Alpha to Omicron: the origin, significant mutation in the genome, and S-glycoprotein
In this study, we have developed different statistical models to depict the evolution of SARS-CoV-2 variants and the data and information collection of these variants. This study has considered significant VOCs and VOIs along with the Wuhan strain. We have considered significant VOCs such as B.1.351 (Beta), B.1.1.7 (Alpha), B.1.617.2 (Delta), P.1 (Gamma), and B.1.1.529 (Omicron). At the same time, we have considered significant VOIs such as B.1.525 (Eta), B.1.621 (Mu), C.37 (Lambda), B.1.526 (Iota), P.2 (Zeta), and B.1.427/B.1.429 (Epsilon), B.1.616, P.3 (Theta).
Continent-wise evolution of SARS-CoV-2 and its variants with their clades and divergence
The NextStrain server was used for the phylogenetic analysis of SARS-CoV-2 [38]. Researchers have used this server to illustrate the phylodynamics of real-time pathogen evolution [39]. Researchers have demonstrated continent-wide analysis to understand the relationship with the epidemiology of COVID-19 and how SARS-CoV-2 evolution can alter mortality, virulence, and infectivity [40]. The NextStrain clades were analyzed continuously in this study, providing information on the continent-wise SARS-CoV-2 evolution. In this part of the study, statistical software (PAST 4.03 software) was used to depict the divergence models of SARS-CoV-2 evolution [41].
Lineage dynamics and comparison between SARS-CoV-2 lineages across different countries
Our study evaluated the lineage dynamics and compared various lineages from different countries. The CoVerage server was used to analyze the lineage dynamics of SARS-CoV-2 [42]. This server acquires the original data from GISAID ([43] and web-based repositories [44].
Gene-wise mutation patterns and genetic variability of SARS-CoV-2 variants across various geographic locations
At this point of the pandemic, it is necessary to evaluate the mutation patterns of different genes of the variants from several countries, which can provide a clue about the mutation patterns and genetic variability of SARS-CoV-2. Thus, the study analyzed the gene-wise mutation patterns and genetic variability of SARS-CoV-2 variants across different geographic locations. For this analysis, the genomic variability and global testing-related server developed by the researchers at Khazen Lab [45] were utilized.
Significant amino acid point mutations in SARS-CoV-2 variants, their frequencies, and country-wise occurrence
Amino acid (AA) point mutations are an important basis for variant creation. Therefore, the study focus on understanding the AA mutation spectrum of SARS-CoV-2 variants. The CovMT server [46] was used to analyze the frequencies of AA point mutations and country-wise occurrences.
Statistical models and software
Statistical models are significant for understanding evolutionary phenomena and bioinformatics models [47, 48]. We developed different statistical models to understand the mutation pattern of significant mutations. The linear regression model was developed to understand the mutation pattern in S-glycoprotein and the genome of different variants (VOCs and VOIs). On the other hand, a linear model of the scatter plot was developed to understand the clustering events. Another statistical model (box-plot) was developed using different variants to illustrate the divergence patterns of SARS-CoV-2 variants in each continent. Statistical software (PAST 4.03 software) was used to develop various statistical models [41]. MATLAB software was used to depict plots, graphs, and pie charts [49].
Results
SARS-CoV-2 variants from Alpha to Omicron: the origin, significant mutation in the genome, and S-glycoprotein
The variants are emerging due to the different mutations and are being classified as VOCs and VOIs of SARS-CoV-2. It has been noted that a significant variant emerges from a country in the world at a particular time. After the origin of the SARS-CoV-2 in Wuhan (China), the first significant variant reported was B.1.351 (Beta), and according to the WHO, it was the first reported variant in South Africa (Fig. 1a), documented in May 2020 (Fig. 1b). Similarly, B.1.1.7 (Alpha) was reported in the UK in September 2020. It was subsequently transmitted worldwide [50]. Similarly, another significant variant, B.1.617.2 (Delta), originated in India in October 2021 and spread worldwide. Other variants were generated in 2020, such as P1 (first reported in Brazil), B1.526 (registered early in the UK), and C.37 (reported early in Peru). However, in the following year, B.1.621 was reported in early 2021 in the UK. At the same time, P3 (originating in Brazil) and two others (B1.427, B1.429) originated from the USA. In late 2021, Omicron (B.1.1 529) arose from South Africa (Fig. 1a and b) and subsequently spread worldwide. This variant has been reported to have the highest number of mutations with rapid transmissibility. Infectivity is also noted to be high in Omicron, which caused a rise in infection cases of COVID-19 in every country [51]. Omicron exhibits substantial nAb escape properties [52, 53].
SARS-CoV-2 variants and origin, the significant mutation in whole-genome and S-glycoprotein and developed linear model. (a) Schematic diagram shows the country of origin of emerging SARS-CoV-2 variants along with the Wuhan strain. (b) Different time points of origin of emerging SARS-CoV-2 variants. (c) Schematic diagram shows significant mutations in the whole genome of VOCs, excluding S-glycoprotein. (d) Schematic diagram shows significant mutations in the whole genome of VOIs, excluding S-glycoprotein. (e) Schematic diagram shows significant mutations in S-glycoprotein of VOCs. (f) Schematic diagram shows significant mutations in S-glycoprotein of VOIs. (g) Linear model depicted using significant mutations in S-glycoprotein. (h) Linear model illustrated using significant mutations in the whole-genome excluding S-glycoprotein. (i) Linear model depicted using significant mutations in the whole-genome. In this diagram, we used red and green points. The red color indicates VOCs, and the green color indicates VOIs
Mutations are necessary elements to calculate the different evolutionary parameters, such as mutation rate and the molecular clock [54,55,56]. Our study shows significant mutations in the SARS-CoV-2 variants throughout the genomes of VOCs (Fig. 1c) and VOIs (Fig. 1d). The two figures (Fig. 1c and d) illustrate the mutations throughout the genomes of VOCs (Fig. 1c) and VOIs (Fig. 1d) other than the S-glycoprotein mutations.
It was found that point mutation P314L was prevalent at high frequencies in ORF1b in most variants. Hemachudha et al. (2022) also reported the presence of the mutation from the genome sequences of five infected individuals and observed that P314L influences binding affinity to Favipiravir. In addition, it may also augment the binding affinity to remdesivir [57]. Furthermore, researchers have reported from the whole genome surveillance of SARS-CoV-2 from Mexican patients that the mutation P314L was found in ORF1b with single clustering variants [58]. Additionally, we have illustrated significant mutations in the S-glycoprotein of VOCs (Fig. 1e) and VOIs (Fig. 1f). These two figures illustrate the mutations throughout the S-glycoprotein of VOCs (Fig. 1e) and VOIs (Fig. 1f). Figures display the mutations in very detail in every region of the S-glycoprotein.
Several researchers have reported E484K, K417T/N, N501Y, L452R, D614G, and P681R mutations, and these mutations are common in VOIs and VOCs [7, 8, 59,60,61]. The N501Y mutation in S-glycoprotein helps increase the transmission and infection rate of SARS-CoV-2 [62]. Similarly, E484K and N501Y mutations in S-glycoprotein decrease the affinity for nAbs [8]. Barton et al. (2021) noted that the five RBD mutations, S477N, E484K, N501Y, K417T, and K417N, may affect binding affinity to ACE2 receptor [63]. The D614G mutation has also been reported in all VOCs and VOIs, and this mutation enhances the receptor-binding affinity. It has been reported to augment infectivity [3, 10, 64].
Mutational load is a significant factor for RNA viruses and is responsible for the stochastic evolutionary result [65]. Researchers have applied the linear regression model to understand the changes in AA mutations and correlate them with antigenic variants of the influenza virus [66].
We next developed the statistical model to understand the mutation pattern in all significant variants. A linear regression model was developed to depict the mutational load (in terms of the number of mutations) of VOIs and VOCs. A model was designed to illustrate the mutational load throughout S-glycoprotein by utilizing the number of mutations (VOCs and VOIs). It resulted in the determination of the coefficient (R2 = 0.0911) of the linear model (Fig. 1g). This linear model showed that the point representing the Omicron is not close to the regression line (Fig. 1g). Another linear model was developed to depict the mutational load throughout the genome other than the S-glycoprotein using the number of mutations of VOCs and VOIs. Here, we considered the number of mutations throughout the genome, excluding S-glycoprotein. This enabled the determination of the coefficient (R2 = 0.1874) of the linear model (Fig. 1h). Similarly, an additional linear model depicting the mutational load throughout the genome of SARS-CoV-2 (R2 = 0.1426) was evaluated (Fig. 1i). This statistical model considered the mutational load throughout the genome of significant VOCs and VOIs.
Continent-wise evolution of SARS-CoV-2 and its variants with their clades and divergence
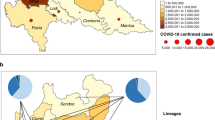
The continental evolution of SARS-CoV-2 variants was studied to understand each continent’s evolution pattern. The continent-wise evolution of SARS-CoV-2 variants was analyzed, including that of African (Fig. 1A (A1 to A3)), Asian (Fig. 1B (B1 to B3)), European (Fig. 1C (C1 to C3)), North American (Fig. 1D (D1 to D3)), South America (Fig. 1E (E1 to E3)), and Oceanian (Fig. 1F (F1 to F3)) variants with their clades and divergence. In Africa, a radial-type polygenetic tree was developed to understand the phylodynamics of the SARS-CoV-2 variants. It was found that Delta was dominant in the phylogenetic tree with three clades (21A, 21I, and 21J) compared to the other variants (Fig. 2a (A1)). However, the 21J clade was more prominent than the two other Delta clades. In Delta, the maximum divergence was noted as 49, and the minimum was 35 (Table 1). It was observed that two clades were developed for Omicron (21L and 21K). Noteworthy, 21K was the major clade among these two clades. A very low presence of Alpha (20I) and Beta (20H) variants was found on this continent. Furthermore, a very low amount of Eta (21D) was also found. A linear model of the scatter plot was developed with a regression line (R2 = 0.779) (Fig. 2a(A2)). In this model, the dots representing Omicron are located at the upper side of the regression line. Furthermore, a scatter plot was developed using the sampling date. The scatter plot showed the initial coexistence of Delta and Omicron. Finally, the Omicron variant was found significant, as shown by this continent’s scatter plot (Fig. 2a(A3)).
Region-wise evolution of SARS-CoV-2 and their variants with their clades and divergence. (a) Evolution of SARS-CoV-2 in Africa. (a1) Radial type of polygenetic tree demonstrates the evolution of variants in Africa. (a2) Scatter plot with a regression model using the variants in Africa. (a3) Scatter plot with sampling date in Africa. (b) Evolution of SARS-CoV-2 in Asia. (b1) Radial type of polygenetic tree demonstrates the evolution of variants in Asia. (b2) Scatter plot with a regression model using the variants in Asia. (b3) Scatter plot with sampling date in Asia. (c) Evolution of SARS-CoV-2 in Europe. (c1) Radial type of polygenetic tree demonstrates the evolution of variants in Europe. (c2) Scatter plot with a regression model using the variants in Europe. (c3) Scatter plot with sampling date in Europe. (d) Evolution of SARS-CoV-2 in North America. (d1) Radial type of polygenetic tree demonstrates the evolution of variants in North America. (d2) Scatter plot with a regression model using the variants in North America. (d3) Scatter plot with sampling date in North America. (e) Evolution of SARS-CoV-2 in South America. (e1) Radial type of polygenetic tree demonstrates the evolution of variants in South America. (e2) Scatter plot with a regression model using the variants in South America. (e3) Scatter plot with sampling date in South America. (f) Evolution of SARS-CoV-2 in Oceania. (f1) Radial type of polygenetic tree demonstrates the evolution of variants in Oceania. (f2) Scatter plot with a regression model using the variants in Oceania. (f3) Scatter plot with sampling date in Oceania. (g) A Box plot-based model illustrates the minimum and maximum divergence event of SARS-CoV-2 variants with their clade in different continents. (g1) A model for divergence event in Africa with (g2) A model for divergence event in Asia, (g3) A model for divergence event in Europe, (g4) A model for divergence event in North America, (g5) A model for divergence event in South America, (g6) A model for divergence event in Oceania
The study next analyzed the evolution of SARS-CoV-2 in Asia. A radial polygenetic tree was developed to understand SARS-CoV-2 evolution in Asia. It was found that Delta was a significant variant in the phylogenetic tree with three clades (21B, 21I, and 21J) (Fig. 2b(B1)). A maximum divergence of 57 was observed, and the minimum was 54 for Omicron (Table 1). In addition, a linear model of the scatter plot was developed with a regression line (R2 = 0.779), and two clusters of Omicron were observed (Fig. 2b(B2)). In this model, the dots representing Omicron are present on the upper side of the regression line, and Alpha and Beta are located in the middle. In addition, a scatter plot was developed using the sampling date (Fig. 2b(B3)). The scatter plot showed a linear cluster with more dots representing Delta in the second plot compared to the African continent.
To understand the evolution of SARS-CoV-2 in Europe, the developed radial polygenetic tree informed the creation of different Beta, Alpha, Omicron, and Delta variants with their clades (Fig. 2c(C1)). Beta and Alpha formed a small cluster in the polygenetic tree. Additionally, a linear model with a regression line was developed, and two clusters of Omicron were found (Fig. 2c(C2)). However, an upper cluster of Omicron was found in this model, which is a very small cluster compared to the Asian continent region. Two different clades formed these two clusters. Among them, the larger cluster contains the variants of 21K, and the smaller cluster includes the variants of 21L. Another scatter plot showed that the presence of the 21A clade of Delta is significantly smaller than the 21I and 21J clades of Delta (Fig. 2c(C3)). The analysis also revealed that the 21K clade of Omicron is prevalent compared to the 21L clade.
For further understanding, SARS-CoV-2 evolution in North America was studied. Phylogenetic analysis showed that the Lota and Gamma variants originated with the Alpha variant (Fig. 2d(d1)). The linear model with a regression line showed a new Omicron clade (21M), which was present below the line (Fig. 2d(d2)). The model showed a larger cluster of Omicron arranged above the regression line. Another model of the scatter plot showed two clades of Omicron (21M and 21K) and three clades of Delta (21I, 21J, and 21K) (Fig. 2d(d3)).
SARS-CoV-2 evolution was illustrated in South America through polygenetic analysis of SARS-CoV-2 variants, demonstrating three prominent parts of the tree: one part of the tree was composed of Gamma, Alpha, Lambda, and Mu variants; another part of the tree consisted of Omicron; and Delta was organized in another part of the tree (Fig. 2e(e1)). It was observed from the developed linear model with the regression line that Alpha and Omicron were arranged above the regression line. Conversely, the dots of Delta were mostly arranged below the line (Fig. 2e(e2)). In this model, significantly fewer dots were noted for Mu and Lambda. Additionally, another scatter plot was developed with the sampling date (Fig. 2e(e3)). The scatter plot showed a single line of Omicron dots that were arranged very closely. Limited Mu and Lambda dots were noted, arranged in two lines and with detached dots.
In Oceania, the radial polygenetic tree of SARS-CoV-2 informed that Kappa (21B) had the same point of origin with two Delta clades (21A and 21I) and formed a cluster with these variants. The phylogenetic analysis showed that the number of analyzed sequences of the Delta variant (clade 21J) is less compared to other continents (Fig. 2f(f1)). The linear model with a regression line showed two Omicron clades (21L and 21K) arranged above the bar. Similarly, the 21J Delta clade was mainly placed below the line (Fig. 2f(f2)), and the 21A clade of Delta is noted to be very less in number and arranged below the line. The model showed the two clusters of Omicron placed above the regression line. Another model of the scatter plot showed two clades of Omicron (21K, 21L). More Omicron dots were found in clade 21K compared to clade 21L. A higher number of dots were found in the clade 21J Delta variants, corresponding to other clades of the Delta variant (21A, 21I) (Fig. 2f(f3).
Divergence has been used to understand microbial diversity (Lozupone and Knight, 2008)[93]. The virus variants and their clades can be elucidated through divergence. To understand the divergence event, Boni et al. (2020) studied divergence dates between the bat Sarbecovirus and SARS-CoV-2 [67]. Divergence represents the splitting event of a variant into a phylogenetic tree. Tang et al. (2020) analyzed the molecular divergence of Coronaviruses to understand the connection between SARS-CoV-2 and other associated Coronaviruses [13]. In this study, a statistical model was developed to understand the divergence patterns of SARS-CoV-2 variants. The model showed the divergence events of SARS-CoV-2 variants in different continents such as Africa (Fig. 2g(g1)), Asia (Fig. 2g(g2)), Europe (Fig. 2g(g3)), North America (Fig. 2g(g4)), South America (Fig. 2g(g5)), and Oceania (Fig. 2g(g5)). The model observed the highest divergence event in Omicron (clade 21L) in Asia. The lowest divergence event was noted in Iota (clade 20F) in Asia and North America. However, a maximum divergence event was found in Omicron in Asia and Africa. In both cases, the leading divergence event was recorded in clade 21L of Omicron (Table 1).
Finally, a substantial clade was identified for a specific variant from a particular continent (Table 2). One example of a substantial clade is 21K in Omicron, which is prominent among the two clades (21L and 21K) in the African continent.
Lineage dynamics and comparison between SARS-CoV-2 lineages across different countries
Lineage dynamics provide information on the transmission and dissemination pattern prototype of the virus variants. Researchers have aimed to comprehend the SARS-CoV-2 dissemination pattern in a particular country, and it can illustrate the spreading pattern of a particular variant of SARS-CoV-2 in a country or the emergence of different variants of SARS-CoV-2 in specific countries. This type of study is essential for understanding and gaining knowledge of the super-spreading events of the variants and their transmission properties, which is also associated with genomic epidemiology studies [68]. This analysis is necessary for disease control, and Resende et al. (2021) illustrated the dissemination pattern and evolutionary dynamics of a particular linage (B.1.1.33) of SARS-CoV-2 in Brazil [69]. Similarly, Jha et al. (2021) illustrated the transmission dynamics of different variants of SARS-CoV-2 across India and suggested that it may be helpful for strategy-making for public health preparedness and decision-making in several parts of India [70].
Our study attempted to understand lineage dynamics and compare different lineages from different countries. From this perspective, a heatmap was generated to compare the lineages across regions (Fig. 3a). This map represents the country-wise lineage frequency of the virus, showing the country-wise spread of the different lineages of SARS-CoV-2. Moreover, country-wise lineage dynamics were analyzed using a country-wise lineage dynamics plot to illustrate the rise and fall of the variants over the years. At first, a lineage dynamics plot of Germany was developed, which illustrated the increase in significant lineages such as B.1.329 (from April to August 2020), B.1.1.7 (Alpha) (from December 2020 to July 2021), B.1.617.2 (Delta) (from May 2021 to January 2022 or so), and B.1.1 529 (Omicron) (from November 2021 onwards) among all SARS-CoV-2 lineages (Fig. 3b). The lineage dynamics plot of the UK illustrated the rise of B.1.177 (from July 2020 to March 2021), B.1.1.7 (Alpha) (from October 2020 to June 2021), B.1.617.2 (Delta) (from March 2021 to January 2022 or so), and B.1.1 529 (Omicron) (from November 2021 onwards), which have each dominated in the country over time (Fig. 3c). The developed dynamics plot illustrated the rise of the B.1.1.7 variant (from January to February 2021) in September 2020. Next, a lineage dynamics plot was developed for the USA. This illustrated the augmentation of important lineages such as A.1 (from January to May 2020), B.1.1.7 (Alpha) (from January to July 2021), B.1.429 (Epsilon) (from November 2020 to May 2021), B.1.617.2 (Delta) (from May 2021 to January 2022 or so), and B.1.1 529 (Omicron) (from November 2021 onwards) among all the SARS-CoV-2 lineages (Fig. 3d). It was observed that B.1.429 originated in November 2020 in the USA. A lineage dynamics plot was also developed for France. This illustrated the emergence of important lineages in this country, such as B.1.159 (from May to July 2020), B.1.367 (from June to October 2020), B.1.1.7 (Alpha) (from November 2020 to July 2021), B.1.617.2 (Delta) (from May 2021 to January 2022), AY.4 (from June 2021 to January 2022), BA.1 of B.1.1 529 (Omicron) (from November 2021 onwards) among all the SARS-CoV-2 lineages (Fig. 3e). Similarly, the generated lineage dynamics plot of India illustrated the emergence of B.1.306 (from March to December 2020), B.1.1.326 (from April to November 2020), B.1.1.7 (Alpha) (from December 2020 to May 2021), B.1.617.2 (Delta) (from October 2020 to January 2022 or so), AY.112 (from March 2021 to January 2022), AY.127 (from June 2021 to January 2022), BA.1 and BA.2 of B.1.1 529 (Omicron) (from Decrember 2021 onwards) lineages in the country (Fig. 3f). The dynamics plot illustrated the origin of VOI B.1.617.2 (Delta) during October 2020 and the successive rise of the variant. Furthermore, the lineage dynamics plot of Brazil illustrated SARS-CoV-2 lineage origin and increase in the country, including the P.1 (Gamma) (from November 2020 to October 2021), P.2 (Zeta) (from August 2020 to April 2021), B.1.617.2 (Delta) (from June 2021 to January 2022), AY.43 (from June 2021 to January 2022), BA.1, of B.1.1 529 (Omicron) (from November 2021 onwards), which have each dominated at some time (Fig. 3g). The plot illustrated the origin of VOI P.1 (Gamma) during November 2020 and the rise of another variant, P.2 (Zeta), since April 2020. Next, the lineage dynamics plot of South Africa illustrated the spread of lineages C.1 (from March to November 2020), B.1.54 (from May to December 2020), B.1.351 (Beta) (from August 2020 to July 2021), B.1.617.2 (Delta) (from February to December 2021), AY.45, AY.38, AY.32, BA.1 of B.1.1 529 (Omicron) (from November 2021 onwards) in the country (Fig. 3h). Two lineages, B.1.351 (Beta) and B.1.1 529 (Omicron), originated from this region, and it has been found that B.1.351 (Beta) originated in May 2020 and B.1.1 529 (Omicron) in November 2021. Finally, the study of Singapore illustrated the spread of B.1, B.6.6, B.6.4, B.1.351 (Beta) (from February to May 2021), B.1.617.2 (Delta) (from March 2021 to January 2022 or onwards), AY.23, BA.1 of B.1.1 529 (Omicron) (from November 2021 onwards) in the country. Here, B.1, B.6.6, B.1.617.2 (Delta), and AY.23 variants increased significantly. Finally, we have shown different countries-wise lineages and their time period of dominancy in a table format (Table 3).
Lineage dynamics and comparison between SARS-CoV-2 lineages across different countries (a) a heatmap that compares the lineages across regions. (b) Lineage dynamic plots illustrate the rise of the several lineages in Germany. (c) Lineage dynamic plots illustrate the increase of the several lineages in the UK. (d) Lineage dynamic plots illustrate the rise of the several lineages in the USA. (e) Lineage dynamic plots illustrate the increase of the several lineages in France. (f) Lineage dynamic plots illustrate rise of the several lineages in India. (g) Lineage dynamic plots illustrate augment of the several lineages in Brazil. (h) Lineage dynamic plots illustrate rise of the several lineages in South Africa. (i) Lineage dynamic plots illustrate the increase of the several lineages in Singapore
Gene-wise mutation patterns and genetic variability of SARS-CoV-2 variants across various geographic locations
Genetic variability in SARS-CoV-2 has created the genetic diversity of the virus, which is crucial for its survival, fitness, and pathogenesis [71]. Mutations in different parts of the genome have resulted in the emergence of different variants. Therefore, a study of the mutations would provide new insights into the development of SARS-CoV-2 variants. The mutation rate of the virus until September 2020 was determined to be approximately 8 × 10−4 nucleotides/genome per year [71, 72]. However, the SARS-CoV-2 mutation pattern varies in different geographic locations and at different points of the period [73]. It is essential to study the mutation patterns of different genes of the variants from several countries.
We have studied synonymous and nonsynonymous substitutions in the SARS-CoV-2 genome. Synonymous substitutions, also entitled point mutations in nucleotide, occur in a particular position in the nucleotide, but no change in amino acid level is observed. However, in the case of nonsynonymous mutations, one nucleotide might be deleted or inserted, causing at least one amino acid change at the protein level. Understanding the synonymous and nonsynonymous substitutions in the SARS-CoV-2 genome is highly important, and other researchers have also tried to understand the synonymous and nonsynonymous substitutions in the MERS-CoV, SARS-CoV-1, and SARS-CoV-2 genome [74]. Simultaneously, it is also important to understand the transition and transversions events in the SARS-CoV-2 genome. The interchange event between the purines (from a purine to purine; two-ring purine) (A ↔ G) or between the pyrimidines (from a pyrimidine to pyrimidine; one-ring pyrimidines) (C ↔ T) is entitled as the transition. During transversions, the interchanges are noted between the two rings (purine) to one ring (pyrimidine) or vice versa (A or G ↔ C or T) [75]. Like us, Roy et al. have also tried to understand the transition and transversions events in the SARS-CoV-2 genome. They found 12,594 nos of transitions and 7569 nos of transversions from a total of 20,163 events. They also observed that transitions were predominated by cytidine-to-uridine, and transversions were predominated by guanosine-to-uridine conversions [76].
A genetic variability plot was developed with mutation percentage per gene and different mutation consequence percentages for the SARS-CoV-2 genome. First, the study depicted a genetic variability plot for all genes submitted to GISAID from December 2019 to January 2022 (Fig. 4a). It shows the average mutation/1 kb for all genes and delivered an average of 40 mutations per 1 kb gene. Similarly, the developed mutation percentage per gene showed the maximum mutation percentage in ORF10 (16.92%) and the minimum mutation percentage in the E gene (1.82%) (Fig. 4b). Additionally, the different mutation consequence percentage analysis showed a high occurrence of the missense mutation (approximately 53.62%) and synonymous mutation (approximately 34.48%) (Fig. 4c). A country-wise genetic variability plot was developed with mutation percentage per gene and different mutation consequence percentages for the SARS-CoV-2 genome. A country-wise genetic variability plot developed for the USA showed a maximum in the ORF6 gene (approximately 160 mutations/kb) (Fig. 4d). Moreover, the developed mutation percentage per gene showed the maximum ORF3A (15.18%) and minimum mutation percentage in the E gene (2.17%). The mutation in the S gene was 5.56% in the USA (Fig. 4e). Similarly, the consequence mutation percentage illustrated the high occurrence of missense mutations (approximately 60.31%) and synonymous mutations (approximately 28.27%) in the USA (Fig. 4f). Furthermore, genetic variability in Brazil was plotted, showing maximum being in the ORF6 gene (approximately 130 mutations/kb) (Fig. 4g). The mutation percentage per gene was calculated, illustrating the maximum mutation percentage in ORF6 (27.7%) and the minimum in the E gene (0.93%). The mutation in the S gene was 5.6% in Brazil (Fig. 4h). Similarly, the consequence mutation percentage in Brazil was evaluated, which observed an elevated incidence of missense mutations (about 62.31%) and synonymous mutations (approximately 27.93%) (Fig. 4i).
Gene-wise mutation pattern and genetic variability of SARS-CoV-2 variants across various geographic locations. (a) Genetic variability plot for all genes using the submitted sequence from December 2019 to January 2022. (b) Mutation percentage per gene. (c) Average distribution of different consequences of mutational event. (d) Genetic variability plot for USA. (e) Mutation percentage per gene for USA. (f) Average distribution of different consequences of mutational event for USA. (g) Genetic variability plot for Brazil. (h) Mutation percentage per gene for Brazil. (i) Average distribution of different consequences of mutational event for Brazil. (j) Genetic variability plot for South Africa. (k) Mutation percentage per gene for South Africa. (l) Average distribution of different consequences of mutational event for South Africa. (m) Genetic variability plot for England. (n) Mutation percentage per gene for England. (o) Average distribution of different consequences of mutational event for England
This study also depicted genetic variability in South Africa, where S-gene mutations were more frequent (Fig. 4j). The evaluated mutation percentage per gene in South Africa showed the maximum mutation percentage in the S gene (9.25%) and minimum in the ORF7A gene (2.46%) (Fig. 4k). The consequence mutation percentage in this region showed an elevated incidence of missense mutations, approximately 57.66%, and an incidence of synonymous mutations of approximately 30.68% (Fig. 4l).
Finally, the genetic variability was estimated, showing the consequence of the repeated Sgene mutation event. The genetic variability plot indicated the occurrence of a few cases of maximum mutation in ORF7A (Fig. 4m). The mutation percentage event per gene in the same region was calculated, illustrating the maximum mutation percentage in the ORF10 gene (25.04%) and the minimum in the ORF6 gene (1.63%). A minimum mutation percentage in the E gene (1.87%) was also recorded (Fig. 4n). In England, the mutation outcome percentage showed an increased frequency of missense mutations (approximately 52.44%). The second-highest percentage of synonymous mutations was also observed, which was approximately 38.18% (Fig. 4o).
Significant amino acid point mutations in SARS-CoV-2 variants, their frequencies, and country-wise occurrence
Amino acid (AA) point mutations alter protein structure and function. Researchers have attempted to predict AA point mutations due to their altered structure and function, which have clinical significance [77]. AA point mutations in SARS-CoV-2 variants have immense importance from a clinical perspective [78]. Understanding the AA point mutations in SARS-CoV-2 variants will help comprehend the variant’s pathogenicity. These mutations result in more resistant variants [78]. The assessment of these AA point mutations is vital in studying SARS-CoV-2 drug resistance, immune escape, antibody escape, and partial vaccine escape events [79].
A heatmap of AA point mutations with cumulative and frequency analyses of different point mutations (T478K, L452R, N501Y, S477N, E484A, Q498R, and Y505H) and the occurrence of these mutations in several countries was generated. The generated heatmap of AA point mutations illustrated the highest event of two mutations (T478K and L452R) (Fig. 5a). The cumulative occurrence, frequency, and country-wise occurrence of T478K were analyzed. Two types of cumulative graphs were generated for T478K, which were cumulative analyses using monthly data (Fig. 5b(b1)) and weekly data (Fig. 5b(b2)), displaying the mutation frequency increase from July 2021. Two types of frequency analysis graphs for T478K were also generated: frequency analysis using monthly data (Fig. 5b(b3)) and weekly data (Fig. 5b(b4)), showing that the mutation frequency increased from July 2021 and decreased at the end of 2021. The heatmap of T478K is also depicted to understand the country-wise occurrence of this mutation, and it illustrated the highest number of T478K in Senegal during September 2021 (Fig. 5b(b5)).
Significant AA point mutations in SARS-CoV-2 variants, their frequencies, and country-wise occurrence. (a) Heatmap of AA point mutations. (b) Cumulative and frequency analysis of T478K point mutations: (b1) cumulative graph of T478K mutation using monthly data; (b2) cumulative graph of T478K mutation using monthly data; (b3) frequency graph of T478K mutation using monthly data; (b4) frequency graph of T478K mutation using monthly data; (b5) a heatmap illustrates the country-wise occurrence of T478K mutation. (c) Cumulative and frequency analysis of L452R point mutations: (c1) cumulative graph of L452R mutation using monthly data; (c2) cumulative graph of L452R mutation using monthly data; (c3) frequency graph of L452R mutation using monthly data; (c4) frequency graph of L452R mutation using monthly data; (c5) a heatmap illustrates the country-wise occurrence of L452R mutation; (d) Cumulative and frequency analysis of N501Y point mutations; (d1) cumulative graph of N501Y mutation using monthly data; (d2) cumulative graph of N501Y mutation using monthly data; (d3) frequency graph of N501Y mutation using monthly data; (d4) frequency graph of N501Y mutation using monthly data; (d5) a heatmap illustrates the country-wise occurrence of N501Y mutation. (e) Cumulative and frequency analysis of S477N point mutations: (e1) cumulative graph of S477N mutation using monthly data; (e2) cumulative graph of S477N mutation using monthly data; (e3) frequency graph of S477N mutation using monthly data; (e4) frequency graph of S477N mutation using monthly data; (e5) a heatmap illustrates the country-wise occurrence of S477N mutation. (f) Cumulative and frequency analysis of E484A point mutations: (f1) cumulative graph of E484A mutation using monthly data; (f2) cumulative graph of E484A mutation using monthly data; (f3) frequency graph of E484A mutation using monthly data; (f4) frequency graph of E484A mutation using monthly data; (f5) a heatmap illustrates the country-wise occurrence of E484A mutation. (g) Cumulative and frequency analysis of Q498R point mutations: (g1) cumulative graph of S477N mutation using monthly data; (g2) cumulative graph of Q498R mutation using monthly data; (g3) frequency graph of Q498R mutation using monthly data; (g4) frequency graph of Q498R mutation using monthly data; (g5) a heatmap illustrates the country-wise occurrence of Q498R mutation. (h) Cumulative and frequency analysis of Y505H point mutations: (h1) cumulative graph of Y505H mutation using monthly data; (h2) cumulative graph of Y505H mutation using monthly data; (h3) frequency graph of S477N mutation using monthly data; (h4) frequency graph of Y505H mutation using monthly data; (e5) a heatmap illustrates the country-wise occurrence of Y505H mutation
The cumulative occurrence, frequency, and country-wise occurrence of L452R were also evaluated. Two types of cumulative analysis were illustrated for this point mutation, a monthly cumulative analysis (Fig. 5c(c1)) and a weekly cumulative analysis (Fig. 5c(c2)), which showed that the mutation increased from July 2021. Simultaneously, two types of frequency analysis graphs were generated for L452R on a monthly (Fig. 5c(c3)) and weekly basis (Fig. 5c(c4)). This showed that the mutation frequency increased from July 2021 and decreased at the end of 2021. The heatmap of L452R is also depicted, showing the country-wise occurrence of this mutation, and it illustrated the highest number of L452R in the USA during September 2021 (Fig. 5c (c5)).
The analysis of the N501Y mutation is depicted in the cumulative graphs (cumulative analysis monthly (Fig. 5d(d1)) and cumulative analysis weekly (Fig. 5d(d2)) and frequency analysis graphs (monthly basis (Fig. 5d(d3)), and weekly basis (Fig. 5d(d4)). The mutation frequency increased from January 2021 and decreased in September 2021. However, it was noted that the mutation increased after a few months. The heatmap of N501Y showed this mutation’s country-wise incidence, and the maximum number was found in Senegal and Spain from March 2021 to December 2021 (Fig. 5d(d5)).
The analysis of S477N was performed, and the cumulative graphs (cumulative monthly analysis (Fig. 5e(e1)) and cumulative weekly analysis (Fig. 5e(e2)) and the frequency analysis graphs (monthly (Fig. 5e(e3)) and weekly (Fig. 5e(e4)) are displayed. The analysis revealed that the mutation frequency increased from November 2021 and decreased during 2021. However, it was noted that the mutation increased after a few months. The heatmap of S477N depicted the maximum number in Senegal and France in December 2021 (Fig. 5f(f5)). Similarly, the cumulative graphs (monthly (Fig. 5f(f1) and weekly (Fig. 5f(f2)) and frequency graphs (monthly (Fig. 5f(f3) and weekly (Fig. 5f(f4)) of E484A are depicted. The analysis showed an increase in the E484A mutation after October 2021. The heatmap of E484A depicted the maximum number in Italy during December 2021 (Fig. 5f(f5)).
We next analyzed the cumulative occurrence (monthly (Fig. 5g(g1) and weekly (Fig. 5g(g2)) and frequency of the AA point mutation (Q498R) (monthly (Fig. 5g(g3) and weekly (Fig. 5g(g4)) of E484A. It was found that an increase in the Q498R mutation occurred in December 2021. The generated heatmap of Q498R illustrated the maximum number of mutations in December 2021 (Fig. 5g(g5)). Finally, the present study evaluated the cumulative incidence of Y505H (monthly (Fig. 5h(h1) and weekly (Fig. 5h(h2)) and frequency of Y505H mutation (monthly (Fig. 5h(h3) and weekly (Fig. 5h(h4)) for the UK. It was an augment of the Y505H mutation during December 2021, and after that, the analysis showed an immediate decrease. Again, the generated heatmap of Q498R illustrated the maximum number of mutations in December 2021 in the UK (Fig. 5h(h5)).
Discussion
The current study applied several statistical and bioinformatics models to predict the evolution pattern and mutation prototype of SARS-CoV-2 and its emerging variants. First, three statistical models were developed using different variants to illustrate the significant mutational load throughout S-glycoprotein (Fig. 1g), the whole genome excluding S-glycoprotein (Fig. 1h), and the whole genome. It showed the overall mutation load pattern in S-glycoprotein and throughout the genome of all SARS-CoV-2 variants. The model revealed that significant VOCs and VOIs form a cluster in mutation load in S-glycoprotein and throughout the genome, excluding the Omicron variant. Omicron exhibited a different pattern in terms of mutation load, which was high, and the natural selection may create an increased mutational load for its survival. These mutations might be responsible for increased transmission among the host and enable them to escape from nAbs. Therefore, mutations in Omicron increase “viral fitness” and provide better survival advantages. This may be a process of the adaptive evolution of the SARS-CoV-2 variant [80]. Additionally, the statistical model of continent-wide divergence indicated the maximum divergence event in Omicron, especially in Asia and Africa (Fig. 2g).
Our analysis illustrated the evolution of different SARS-CoV-2 variants in various continents. The analysis shows a typical evolutionary pattern of various variants on multiple continents. However, the phylogenetic patterns and the different factors of evolution of transmission and molecular adaptation are yet to be fully understood [81]. Fischer et al. (2021) performed a comparative analysis of two viruses (SARS-CoV-2 and HIV-1) in terms of structural comparisons of S-glycoprotein and Env proteins, recombination, phylogenetic events, and transition frequency and depicted the evolutionary pattern of the two viruses [82]. Similarly, our analysis revealed lineage dynamics and a comparison between SARS-CoV-2 lineages across different countries and illustrated clusters among clades in a particular variant. Our study also found a general pattern of variant shifts. Variant shifts are prevalent during SARS-CoV-2 evolution, such as Alpha to Delta and Delta to Omicron. It has been noted that significant variants have evolved from time to time through natural selection. Furthermore, it was also observed that the newly naturally selected variant is transmitted quickly and replaces the previous dominant variant through a variant shift. The dominance of the Delta variant has been previously observed [83].
Our analysis of region-wise genetic variability and mutation profiling showed a general trend in the high mutation percentage in selected genes of SARS-CoV-2, such as ORF10, ORF6, and S. These mutations assist structural and functional changes, such as nAb resistance and enable immune escape and transmission to a particular continent population, increase in “viral fitness,” and finally becoming a dominant variant. Researchers have observed the same phenomenon in other studies [84]. A low mutation percentage of the E gene might be responsible for envelope stability.
We observed a high percentage of missense and synonymous mutations in variants from several countries. Our study has illustrated synonymous and nonsynonymous substitutions in the SARS-CoV-2 genome. The single nucleotide substitutions (SNS) are of prime importance in evolution due to the ability to change an amino acid sequence which finally changes the protein level. A study by Abdullah et al. developed a probability matrix that can calculate alteration in AA residues in a protein sequence due to the SNS [85]. Kryazhimskiy et al. have tried to map the dynamics of the synonymous and nonsynonymous nucleotide framework in genes of influenza A virus and observed the different patterns of nucleotide substitution in the internal (deep) branches of the phylogeny. It suggests that the synonymous substitution is affected by natural selection. Finally, they found a high correlation between the nucleotide composition dynamics in the synonymous and nonsynonymous sites [86]. It has been observed that synonymous nucleotide substitutions can be targeted by natural selection at the RNA or DNA level of the RNA or DNA viruses. Cuevas et al. have demonstrated that the selection is stronger at synonymous sites in RNA viruses compared to the DNA viruses. At the same time, the consequence of nonsynonymous substitutions is similar in both the cases of ss (single-stranded) RNA and ss DNA viruses [87].
To comprehend the mutation pattern, understanding the transition and transversions events is essential (Krasnov et al., 2015). We studied transition and transversions events in the SARS-CoV-2 genome. In some cases, it has been observed that substitution rates of transitions are superior compared to transversions. Lyons and Lauring have studied transitions and transversion in two RNA viruses (influenza and HIV) and found the overall substitution of transitions and transversion ratio [88]. Similarly, Matyášek and Kovařík analyzed mutation patterns of Coronavirus (human SARS-CoV-2 and bat RaTG13 CoV) from their genome sequence analysis and found that mutations are biased towards C > U transitions [89].
Researchers have illustrated that spike mutations such as N501Y, E484K, L452R, and P681R increase viral fitness [84, 90]. Our study analyzed the frequency of different amino acid point mutations (T478K, L452R, N501Y, S477N, E484A, Q498R, and Y505H) and heatmap analysis of country-wise occurrence, demonstrating the occurrence of the highest event of two mutations (T478K and L452R). These two mutations followed a general pattern of increase and decrease (an increase from July 2021 and a reduction at the end of 2021). Heatmaps of the different mutations from different countries illustrate their high occurrence in various countries. The N501Y mutation showed an increased transmission rate through better binding with ACE2 [91] and was found in Senegal and Spain, with high occurrence from March 2021 to December 2021.
The evolution of the virus occurs because of mutations and natural selection of the variants. This study has tried to map the continent-wide evolution pattern of the emerging SARS-CoV-2 variants considering significant VOCs and VOIs. In the present study, we also analyzed the significant AA point mutations in SARS-CoV-2 variants, their frequencies, and country-wise occurrence. Previously, we have explained one significant mutation in the SARS-CoV-2 S-glycoprotein (D614G) and how this mutation helps in natural selection [6, 10]. This mutation is noted in the S-glycoprotein of all VOIs and VOCs. The study also focused on the mutations analysis in SARS-CoV-2 variants to understand the continent-wide evolution pattern. Finally, our detailed study illustrated the continent-wide evolution pattern of SARS-CoV-2 variants, assisting in understanding the ongoing process of SARS-CoV-2 evolution.
Conclusion and future prospective
The emergence of SARS-CoV-2 variants with recurrent mutations (increasing “viral fitness”) assists in quick transmission. These events have made the pandemic more critical. Our study developed statistical models for mutation load and divergence events, allowing the researchers to understand the evolution further. Our analysis showed the different variants with their significant clades, the range of divergence, and divergence patterns. The analysis of varying AA point mutations helped depict the frequency increase or decrease trend of a particular mutation at different times. Data will assist future researchers in evaluating the mutations and, simultaneously, the ranking of the mutations can be allocated concerning increase or decrease tendency in the different time frames. These findings provide a deeper understanding of the epidemiological and clinical events such as transmission, immune escape related to variants, and mutations.
Overall, the charge calculation of the mutated residues in the genes of the envelope proteins of SARS-CoV-2 is a fascinating area of research. Very few researchers tried to describe some mutations and amino acid changes and their changes in electric charge [27, 92]. However, it is necessary to evaluate the amino acid changes and their changes in electric charge. We appeal to future researchers to illustrate the charge in the genes and electric charge of the mutated residues to the envelope proteins and all variants of the SARS-Cov-2 (from Alpha to Omicron).
However, our findings provide a deeper understanding of the epidemiological and clinical events such as transmission, immune escape related to variants, and mutations. Interestingly, there are many epidemiological differences from country to country. The reasons are not known. However, some of the reasons for the differences: e.g., different public health measures implemented in a time shift in different countries, high prevalence of other infectious diseases (e.g., HIV infection in South Africa), frequent comorbidities in different age groups, other social factors, etc. However, our generated data might help future researchers to explain more about the epidemiological and clinical events.
Further research should aim to discover the molecular catalyst of mutations and hotspots of these mutants to unfold the epidemiological, pathophysiological, and clinical events. During the present study, several clusters of variants and their clades that formed during the evolution of SARS-CoV-2 were observed, suggesting several clustering events during evolution. However, gene transfer events among these virions in a cluster need to be elucidated, which may solve the molecular determinants of the evolution of SARS-CoV-2. These indications will assist future researchers in unfolding answers about the generation of the SARS-CoV-2 variant, and it might also provide answers for creating a subsequent SARS-CoV-2 variant beyond the Omicron.
Abbreviations
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- COVID-19:
-
Coronavirus disease 2019
- VOC:
-
Variants of concern
- VOI:
-
Variants of interest
- RBD:
-
Receptor-binding domain
- ACE2:
-
Angiotensin-converting enzyme 2
- WHO:
-
World Health Organization
- CDC:
-
Centers for Disease Control and Prevention
- eCDC:
-
European Centre for Disease Prevention and Control
- UK:
-
United Kingdom
- USA:
-
United States of America
- AA:
-
Amino acid
- S-glycoprotein:
-
Spike glycoprotein
- GISAID:
-
Global initiative on sharing avian flu data
- nAbs:
-
Neutralizing antibodies
References
Lauring AS, Hodcroft EB. Genetic variants of SARS-CoV-2—what do they mean? JAMA. 2021;325:529–31.
Oude Munnink BB, Worp N, Nieuwenhuijse DF, Sikkema RS, Haagmans B, Fouchier RA, et al. The next phase of SARS-CoV-2 surveillance: real-time molecular epidemiology. Nat Med. 2021;27:1518–24.
Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(812–27):e19.
Trucchi E, Gratton P, Mafessoni F, Motta S, Cicconardi F, Mancia F, et al. Population dynamics and structural effects at short and long range support the hypothesis of the selective advantage of the G614 SARS-CoV-2 spike variant. Mol Biol Evol. 2021;38:1966–79.
Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11:1–9.
Bhattacharya M, Chatterjee S, Sharma AR, Agoramoorthy G, Chakraborty C. D614G mutation and SARS-CoV-2: impact on S-protein structure, function, infectivity, and immunity. Appl Microbiol Biotechnol. 2021;105:9035–45.
Chakraborty C, Bhattacharya M, Sharma AR. Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: their significant mutations in S‐glycoprotein, infectivity, re‐infectivity, immune escape and vaccines activity. Rev Med Virol. 2021: e2270.
Chakraborty C, Bhattacharya M, Sharma AR. Emerging mutations in the SARS-CoV-2 variants and their role in antibody escape to small molecule-based therapeutic resistance. Curr Opin Pharmacol. 2022;62:64–73.
Chakraborty C, Bhattacharya M, Sharma AR, Lee S-S, Agoramoorthy G. SARS-CoV-2 Brazil variant in Latin America: more serious research urgently needed on public health and vaccine protection. Ann Med Surg. 2021: 102428.
Chakraborty C, Saha A, Sharma AR, Bhattacharya M, Lee S-S, Agoramoorthy G. D614G mutation eventuates in all VOI and VOC in SARS-CoV-2: Is it part of the positive selection pioneered by Darwin? : Elsevier; 2021. p. 237–41.
Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci. 2020;117:9241–3.
Li T, Liu D, Yang Y, Guo J, Feng Y, Zhang X, et al. Phylogenetic supertree reveals detailed evolution of SARS-CoV-2. Sci Rep. 2020;10:1–9.
Tang X, Wu C, Li X, Song Y, Yao X, Wu X, et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020;7:1012–23.
Zehender G, Lai A, Bergna A, Meroni L, Riva A, Balotta C, et al. Genomic characterization and phylogenetic analysis of SARS-COV-2 in Italy. J Med Virol. 2020;92:1637–40.
Kumar S, Tao Q, Weaver S, Sanderford M, Caraballo-Ortiz MA, Sharma S, et al. An evolutionary portrait of the progenitor SARS-CoV-2 and its dominant offshoots in COVID-19 pandemic. Mol Biol Evol. 2021;38:3046–59.
Islam MR, Hoque MN, Rahman MS, Alam A, Akther M, Puspo JA, et al. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci Rep. 2020;10:1–9.
Rahman MS, Islam MR, Alam ARU, Islam I, Hoque MN, Akter S, et al. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J Med Virol. 2021;93:2177–95.
Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16:e3000003.
van Dorp L, Acman M, Richard D, Shaw LP, Ford CE, Ormond L, et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet Evol. 2020;83:104351.
Van Egeren D, Novokhodko A, Stoddard M, Tran U, Zetter B, Rogers M, et al. Risk of rapid evolutionary escape from biomedical interventions targeting SARS-CoV-2 spike protein. PLoS ONE. 2021;16:e0250780.
De Maio N, Walker CR, Turakhia Y, Lanfear R, Corbett-Detig R, Goldman N. Mutation rates and selection on synonymous mutations in SARS-CoV-2. Genome Biol Evol. 2021;13:evab087.
Lythgoe KA, Gardner A, Pybus OG, Grove J. Short-sighted virus evolution and a germline hypothesis for chronic viral infections. Trends Microbiol. 2017;25:336–48.
McCormick KD, Jacobs JL, Mellors JW. The emerging plasticity of SARS-CoV-2. Science. 2021;371:1306–8.
Hershberg R. Mutation—the engine of evolution: studying mutation and its role in the evolution of bacteria. Cold Spring Harb Perspect Biol. 2015;7:a018077.
Kosuge M, Furusawa-Nishii E, Ito K, Saito Y, Ogasawara K. Point mutation bias in SARS-CoV-2 variants results in increased ability to stimulate inflammatory responses. Sci Rep. 2020;10:1–9.
Miller S, Lee T, Merritt A, Pryce T, Levy A, Speers D. Single-point mutations in the N gene of SARS-CoV-2 adversely impact detection by a commercial dual target diagnostic assay. Microbiology spectrum. 2021;9:e01494-e1521.
Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409–24.
Meng B, Kemp SA, Papa G, Datir R, Ferreira IA, Marelli S, et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B. 1.1. 7. Cell rep. 2021;35:109292.
Rahman MS, Islam MR, Hoque MN, Alam ASMRU, Akther M, Puspo JA, et al. Comprehensive annotations of the mutational spectra of SARS-CoV-2 spike protein: a fast and accurate pipeline. Transboundary emerg dis. 2021;68:1625–38.
Schlick T, Zhu Q, Jain S, Yan S. Structure-altering mutations of the SARS-CoV-2 frameshifting RNA element. Biophys J. 2021;120:1040–53.
Chan J, Oo S, Chor CYT, Yim D, Chan JSK, Harky A. COVID-19 and literature evidence: should we publish anything and everything? Acta Bio Medica: Atenei Parmensis. 2020;91:e2020020.
Zuo X, Chen Y, Ohno-Machado L, Xu H. How do we share data in COVID-19 research? A systematic review of COVID-19 datasets in PubMed Central Articles. Brief Bioinform. 2021;22:800–11.
Farooq RK, Rehman SU, Ashiq M, Siddique N, Ahmad S. Bibliometric analysis of coronavirus disease (COVID-19) literature published in Web of Science 2019–2020. J Fam Community Med. 2021;28:1.
Thoma B, Chan TM. Using Google Scholar to track the scholarly output of research groups. Perspectives on medical education. 2019;8:201–5.
ECDC. Implications of the spread of the SARS-CoV-2 B.1.1.529 variant of concern (Omicron) for the EU/EEA-– first update. ECDC: Stockholm. 2021. . ECDC. 2022. (Access date 27/01/2022)
CDC. Science Brief: Omicron (B.1.1.529) Variant. . 2021. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html (Access date 27/01/2022)
WHO. Update on Omicron. 2021. 2021. https://www.who.int/news/item/28-11-2021-update-on-omicron. (Access date 27/01/2022).
Nextstrain. Nextstrain SARS-CoV-2 resources, on Nextstrain. 2021. https://nextstrain.org/sars-cov-2/ (Access date 25/01/2022).
Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–3.
Hamed SM, Elkhatib WF, Khairalla AS, Noreddin AM. Global dynamics of SARS-CoV-2 clades and their relation to COVID-19 epidemiology. Sci Rep. 2021;11:1–8.
Hammer Ø, Harper DA, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:9.
CoVerage. CoVerage: SARS-CoV-2 lineage dynamics. 2021. https://sarscoverage.org/ (Access date 25–01–2022).
Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance. 2017;22:30494.
Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–4.
Tannous H, Akiki S, Boulos RE, Eid CEK, El Hasbani G, Khoueiry LM, et al. SARS-CoV-2 historical global testing and genomic variability. 2020. 22, 322 (2022). https://doi.org/10.1186/s12879-022-07147-2
Alam I, Radovanovic A, Incitti R, Kamau AA, Alarawi M, Azhar EI, et al. CovMT: an interactive SARS-CoV-2 mutation tracker, with a focus on critical variants. Lancet Infect Dis. 2021;21:602.
Kosiol C, Bofkin L, Whelan S. Phylogenetics by likelihood: evolutionary modeling as a tool for understanding the genome. J Biomed Inform. 2006;39:51–61.
Motta S, Pappalardo F. Mathematical modeling of biological systems. Brief Bioinform. 2013;14:411–22.
MathWorks I. MATLAB, High-performance Numeric Computation and Visualization Software: User's Guide: for UNIX Workstations: MathWorks; 1992.
WHO. Tracking SARS-CoV-2 variants. 2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (Access date 31/01/2022).
Ren S-Y, Wang W-B, Gao R-D, Zhou A-M. Omicron variant (B. 1.1. 529) of SARS-CoV-2: Mutation, infectivity, transmission, and vaccine resistance. World J Clin Cases. 2022;10:1.
Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nat. 2021: 1–7.
Dejnirattisai W, Huo J, Zhou D, Zahradník J, Supasa P, Liu C, et al. SARS-CoV-2 Omicron-B. 1.1. 529 leads to widespread escape from neutralizing antibody responses. Cell. 2022.
Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. J Virol. 2010;84:9733–48.
Sanjuán R, Domingo-Calap P. Mechanisms of viral mutation. Cell Mol Life Sci. 2016;73:4433–48.
Otto SP, Day T, Arino J, Colijn C, Dushoff J, Li M, et al. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr Biol. 2021;31:R918–29.
Hemachudha P, Petcharat S, Ampoot W, Ponpinit T, Paitoonpong L, Hemachudha T. Genetic variations from successive whole genome sequencing during COVID-19 treatment in five individuals. New Microbes and New Infect. 2022: 100950.
Boukadida C, Taboada B, Escalera-Zamudio M, Isa P, Ramírez-González JE, Vazquez-Perez JA, et al. Genomic characterization of SARS-CoV-2 isolated from patients with distinct disease outcomes in Mexico. Microbiol spectr. 2022;10:e01249-e1321.
Chakraborty C, Sharma AR, Bhattacharya M, Agoramoorthy G, Lee S-S. Evolution, mode of transmission, and mutational landscape of newly emerging SARS-CoV-2 variants. MBio. 2021;12:e01140-e1221.
Chakraborty S. E484K and N501Y SARS-CoV 2 spike mutants Increase ACE2 recognition but reduce affinity for neutralizing antibody. Int Immunopharmacol. 2022;102:108424.
Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra. India Microorganisms. 2021;9:1542.
Liu Y, Liu J, Plante KS, Plante JA, Xie X, Zhang X, et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nat. 2021: 1–9.
Barton MI, MacGowan SA, Kutuzov MA, Dushek O, Barton GJ, van der Merwe PA. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. Elife. 2021;10:e70658.
Ozono S, Zhang Y, Ode H, Sano K, Tan TS, Imai K, et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat Commun. 2021;12:1–9.
Zhao L, Abbasi AB, Illingworth CJ. Mutational load causes stochastic evolutionary outcomes in acute RNA viral infection. Virus evol. 2019;5:vez008.
Cui H, Wei X, Huang Y, Hu B, Fang Y, Wang J. Using multiple linear regression and physicochemical changes of amino acid mutations to predict antigenic variants of influenza A/H3N2 viruses. Bio-Med Mater Eng. 2014;24:3729–35.
Boni MF, Lemey P, Jiang X, Lam TT-Y, Perry BW, Castoe TA, et al. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat microbiol. 2020;5:1408–17.
Popa A, Genger J-W, Nicholson MD, Penz T, Schmid D, Aberle SW, et al. Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2. Sci transl med. 2020;12:eabe2555.
Resende PC, Delatorre E, Gräf T, Mir D, Motta FC, Appolinario LR, et al. Evolutionary dynamics and dissemination pattern of the SARS-CoV-2 lineage B. 1.1. 33 during the early pandemic phase in Brazil. Front microbiol. 2021: 3565.
Jha N, Hall D, Kanakan A, Mehta P, Maurya R, Mir Q, et al. Geographical landscape and transmission dynamics of SARS-CoV-2 variants across India: a longitudinal perspective. Front genet. 2021;12:753648.
Rahimi A, Mirzazadeh A, Tavakolpour S. Genetics and genomics of SARS-CoV-2: a review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics. 2021;113:1221–32.
Bano I, Sharif M, Alam S. Genetic drift in the genome of SARS COV-2 and its global health concern. J Med Virol. 2022;94:88–98.
Mercatelli D, Giorgi FM. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol. 2020;11:1800.
Sohpal VK. Comparative study: nonsynonymous and synonymous substitution of SARS-CoV-2, SARS-CoV, and MERS-CoV genome. Genomics Inform. 2021; 19.
Krasnov GS, Dmitriev AA, Kudryavtseva AV, Shargunov AV, Karpov DS, Uroshlev LA, et al. PPLine: an automated pipeline for SNP, SAP, and splice variant detection in the context of proteogenomics. J Proteome Res. 2015;14:3729–37.
Roy C, Mandal SM, Mondal SK, Mukherjee S, Mapder T, Ghosh W, et al. Trends of mutation accumulation across global SARS-CoV-2 genomes: Implications for the evolution of the novel coronavirus. Genomics. 2020;112:5331–42.
Schaefer C, Rost B. Predict impact of single amino acid change upon protein structure. BMC genomics: BioMed Cent; 2012. p. 1–10.
Alkhatib M, Svicher V, Salpini R, Ambrosio FA, Bellocchi MC, Carioti L, et al. SARS-CoV-2 variants and their relevant mutational profiles: Update summer 2021. Microbiol spectr. 2021;9:e01096-e1121.
Chakraborty C, Sharma AR, Bhattacharya M, Lee S-S. A detailed overview of immune escape, antibody escape, partial vaccine escape of SARS-CoV-2 and their emerging variants with escape mutations. Front Immunol. 2022;13:801522.
Hirabara SM, Serdan TD, Gorjao R, Masi LN, Pithon-Curi TC, Covas DT, et al. SARS-COV-2 variants: differences and potential of immune evasion. Front Cell Infect Microbiol. 2022: 1401.
Henquell C, Mirand A, Richter J, Schuffenecker I, Böttiger B, Diedrich S, et al. Phylogenetic patterns of human coxsackievirus B5 arise from population dynamics between two genogroups and reveal evolutionary factors of molecular adaptation and transmission. J Virol. 2013;87:12249–59.
Fischer W, Giorgi EE, Chakraborty S, Nguyen K, Bhattarcharya T, Theiler J, et al. HIV-1 and SARS-CoV-2: patterns in the evolution of two pandemic pathogens. Cell Host Microbe. 2021;29:1093–110.
Chakraborty C, Sharma A, Bhattacharya M, Agoramoorthy G, Lee S. A paradigm shift in the combination changes of SARS-CoV-2 variants and increased spread of delta variant (B. 1.617. 2) across the world. Aging dis. 2022;13:927–42.
Perez-Gomez R. The development of SARS-CoV-2 variants: the gene makes the disease. J dev biol. 2021;9:58.
Abdullah T, Faiza M, Pant P, Akhtar MR, Pant P. An analysis of single nucleotide substitution in genetic codons-probabilities and outcomes. Bioinformation. 2016;12:98.
Kryazhimskiy S, Bazykin GA, Dushoff J. Natural selection for nucleotide usage at synonymous and nonsynonymous sites in influenza A virus genes. J Virol. 2008;82:4938–45.
Cuevas JM, Domingo-Calap P, Sanjuán R. The fitness effects of synonymous mutations in DNA and RNA viruses. Mol Biol Evol. 2012;29:17–20.
Lyons DM, Lauring AS. Evidence for the selective basis of transition-to-transversion substitution bias in two RNA viruses. Mol Biol Evol. 2017;34:3205–15.
Matyášek R, Kovařík A. Mutation patterns of human SARS-CoV-2 and bat RaTG13 coronavirus genomes are strongly biased towards C> U transitions, indicating rapid evolution in their hosts. Genes. 2020;11:761.
Negi SS, Schein CH, Braun W. Regional and temporal coordinated mutation patterns in SARS-CoV-2 spike protein revealed by a clustering and network analysis. Sci Rep. 2022;12:1–10.
Tian F, Tong B, Sun L, Shi S, Zheng B, Wang Z, et al. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife. 2021;10:e69091.
Pawłowski P. SARS-CoV-2 variant Omicron (B. 1.1. 529) is in a rising trend of mutations increasing the positive electric charge in crucial regions of the spike protein S. Acta Biochimica Polonica. 2022;69:263–4.
Lozupone, C.A., and Knight, R. (2008). Species divergence and the measurement of microbial diversity. FEMS microbiology reviews 32: 557–578.
Author information
Authors and Affiliations
Contributions
Chiranjib Chakraborty: conceptualization, methodology, data analysis, writing—original draft, project administration. Manojit Bhattacharya: validation, preparation of tables and figures. Ashish Ranjan Sharma: validation, formal analysis, review and editing of the manuscript. Kuldeep Dhama: review and editing. Sang-Soo Lee: review and validation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Chakraborty, C., Bhattacharya, M., Sharma, A.R. et al. Continent-wide evolutionary trends of emerging SARS-CoV-2 variants: dynamic profiles from Alpha to Omicron. GeroScience 44, 2371–2392 (2022). https://doi.org/10.1007/s11357-022-00619-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11357-022-00619-y