Abstract
Hospital sewage is an ecosystem that facilitates the transfer of antibiotic and heavy metal resistance genes and the interaction of human and environmental bacteria. In this environment, we have detected the presence of 7 KPC-2 and BEL-1 co-producing E. coli isolates of two different clones over a 10-month period in the same hospital. All isolates carried blaKPC-2 and the operon mer on the same IncP plasmid of similar size and an IncN plasmid of different size each clone carrying blaBEL-1. Both IncN-blaBEL-1 plasmids shared a 77 kb region containing blaBEL-1 alongside with fosE, bla OXA-10 and aac(6’)-1b genes in a class 3 integron within a Tn3 transposon. The major IncN plasmid contained in addition a region homolog to P1-like bacteriophage RCS47, including the lytic RepL and lysogenic proteins, but other phage regions were incomplete. The characters such as the temporal persistence in sewage, the absence of colonized patients in the hospital or in the region, the presence of a p1 phage-plasmid fusion and the infrequent class 3 integron as genetic platform would indicate that BEL-1-producing isolates could have been generated in situ by adaptation to human sewage. Part of the microbiota in these discharges could be explained by the interactions of sewage ecosystems and not derive directly from the hospital.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hospital effluents could play a role in the generation and spread of antibiotic-resistant determinants (Korzeniewska and Harnisz 2013). In these effluents, the presence of antibiotic-resistant bacteria from patients is combined with the selective pressure of spilled antibiotics and heavy metals (Hubeny et al. 2021). Many resistant genes are located on mobile genetic elements such as plasmids, transposons, and integrons (Brolund and Sandegren 2016). Generally, spontaneous frequency of plasmid conjugation is low; however, various compounds present in hospital effluents such as disinfectants, heavy metals, human treatments such as anticonvulsants, antiepileptic, and antibiotics, have been reported to promote plasmid conjugation rate (Liu et al. 2022). The concentrations of antibiotics and heavy metals required to maintain plasmid-carrying bacteria are subinhibitory in most cases (Gullberg et al. 2014). In this ecosystem, bacteria of clinical origin adapted to this aquatic environment coexist with environmental bacteria and other types of microorganisms in biofilms that favor genetic exchange and selection with subinhibitory substances. Due to the favorable conditions for genetic exchange, new combinations of species and resistance determinants of clinical or non-clinical origin, as well as mobile elements, can be found in hospital sewages.
BEL-1 (Belgium extended beta-lactamase) is a clavulanic acid-inhibited expanded-spectrum ß-lactamase (ESBL) detected in a Pseudomonas aeruginosa strain isolated from a scrotal swab of a 72-year-old in May 2004 in a Belgian hospital. Sequences identified a novel ESBL distantly related to other Ambler class A ESBLs. The blaBEL-1 gene was found as a gene cassette located in a chromosome-borne class 1 integron structure, named In120, bracketed by Tn1404-type transposon sequences and by P. aeruginosa-specific genes, containing three other gene cassettes (aacA4, aadA5, and smr2) (Poirel et al. 2005). Two other BEL variants have been described in Spain: BEL-2 on the chromosome of P. aeruginosa, with enhanced hydrolytic properties against expanded-spectrum cephalosporins, and BEL-3, with a single amino acid substitution (P160S) with respect to BEL-1 (Poirel et al. 2010)(Juan et al. 2010). This ESBL has been initially identified in P. aeruginosa, but subsequently on a non-conjugative plasmid in a Klebsiella pneumoniae isolate recovered from a patient in a Portuguese intensive care unit in 2009 (Papagiannitsis et al. 2015). BEL-1 has the characteristic of increasing the MIC of cefiderocol and ceftolozane/tazobactam when has been cloned into plasmid pUCp24 (Poirel et al. 2022). Cefiderocol is a novel siderophore cephalosporin with broad-spectrum activity including metallo-betalactamases (Falcone and Tiseo 2022), for which there are currently few therapeutic options (Doi 2019).
Horizontal transmission of this determinant has not been described to date and the appearance of mobile elements that facilitate its propagation among carbapenemase producers is a worrying fact. Additionally, the possibility of plasmid BEL enzymes conferring cefiderocol resistance is unknown. Recently, BEL and KPC co-producing E. coli isolates were detected over a 1-year period in the wastewater of a Spanish Hospital. The aim of this work was to characterize the genetic environments and plasmid carriage of these genes.
Materials and methods
Isolates
In a monitoring of wastewater from several hospitals in southern Spain (Romero-Oraá et al 2019), performed monthly, a total of 254 carbapenemase-producing Enterobacterales were identified over a 12-month period of study in Hospital Puerta del Mar in Cádiz, with 78 (31%) producing KPC-2. Of these, 19 (24%) were E. coli. Seven of these 19 isolates were co-producers of BEL and KPC enzymes and were detected, over the study period, in the months April, May, June, August, and October 2018 and January 2019. The hospital has 667 beds, carries out 150,000 stays/year and has 3 intensive care units, which are a reference for hospitals in the province. Samples (1 l) were collected between 10 and 11 h at the outfall of the hospital collector into the municipal sewer. Briefly, the isolates were recovered as follows: 1 ml of wastewaters, after a 10-min agitation to homogenize, were overnight enriched in peptone broth and then were platted on ChromID CARBA agar. Each different colony was identified by using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF). Antimicrobial susceptibility testing was carried out by disc (Oxoid) diffusion agar method by using EUCAST breakpoints (https://www.eucast.org/clinical_breakpoints, 29 June 2023). Additionally, cefiderocol was also tested by using gradient strip (LiofilChem). Carbapenem resistance screening was performed with carbapenemics discs according to EUCAST guidelines and beta-carba test (Biorad). Carbapenemases were studied with NG Carba 5 lateral flow immunochromatography (NG Biotech), carbapenemases inhibitor discs and by PCR (GeneXpert) and ESBLs by double disc of cephalosporins with and without clavulanic according to EUCAST guidelines.
Plasmid analysis
Plasmid analysis was carried out by two strategies. First, transconjugants containing of the BEL-1 plasmid from the 4 first isolates belonging to ST11685 clone were selected with Escherichia coli J53 in conjugation experiments by using MacConkey 4 mg/l cefotaxime as selective medium. All transconjugants were checked by PCR for blaBEL-1. Plasmid DNA was isolated by the method described by Kieser to check the number of plasmids in each transconjugant. Transconjugants were sequenced by Illumina at a × 300 coverage. Reads were mapped against E. coli J53 and unmapped reads were considered plasmidic and were assembled. Secondly, isolates 56 and 410, belonging each to one of the two clones identified, were sequenced using MinION MK1b device. Plasmid-based replicon typing was carried out by using PlasmidFinder and plasmid MLST of IncN plasmid was assigned by using pMLST 2.0. Relaxase typing was carried out by using MOB-typer (Robertson and Nash 2018). Plasmid sequences from Illumina were compared by using BLASTN with those previously reported in GeneBank (NCBI) and was plotted with the Easyfig program.
Whole genome sequencing
Each E. coli isolate as well as each BEL-1 transconjugants were sequenced using an Illumina MiSeq platform and Nextera Flex DNA sample preparation kit. De novo assemblies were obtained using CLC genomics Workbench software (version 9.5.2). Minimum thresholds for contig size and coverage for E. coli isolates were set at 300 bp and 30 × , respectively, and for transconjugants coverage was set at 300 × . Assemblies were annotated using the tools of the Center for Genomic Epidemiology (https://cge.food.dtu.dk/services/): plasmid incompatibility group with PlasmidFinder 2.1 and antibiotic resistance determinants with ResFinder 4.1 with a threshold of 90% identity. Other annotations were carried out with Rapid Annotation using Subsystem Technology (RAST) server (http://rast.nmpdr.org/) and to determine insertion sequences was used ISfinder v2.0 (https://www-is.biotoul.fr). For identification of plasmid prophage sequence in p410N-BEL-1, the PHAge Search Tool Enhanced Release (PHASTER) (Arndt et al. 2016; Zhou et al. 2011) server was employed.
Moreover, isolates 56 and 410, belonging each to one of the two clones identified, were sequenced using MinION MK1b device (Oxford Nanopore Technologies, Oxford, UK). Libraries were prepared using the rapid barcoding kit (SQK-RBK004) and were loaded onto a R9.4 flow cell (Oxford Nanopore Technologies). Collection of raw electronic signal data and live base-calling was performed using the MinKNOW v1.4.2 (filtering criteria: length, > 1000 bp; quality > 8). Unicycler v4.8 was used to generated an hybrid genome with the MinION long-reads and short-reads from Illumina (Wick et al. 2017). Sequence homology searches (percentage BLASTN identity > 95%) were performed to identify similar plasmids. BLAST Ring Image Generator (BRIG) for plasmid comparisons (Alikhan et al. 2011). The complete nucleotide sequence of all isolates and plasmids were deposited publicly in NCBI under BioProject no. PRJNA1035043 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA1035043?reviewer=jnoge5hv437qmjq7a42svid4tm).
Typing
In silico analysis of MLST of the 7 isolates was implemented by MLST 2.0 available on the CGE website (identity = 100% and coverage = 100%). The Enterobase E. coli cgMLST scheme was employed to acquire the cgMLST type based on 2513 target loci of E. coli genomic sequences (Zhou et al. 2020) and Clermont’s phylogroup was assigned with phylotype experiment from Enterobase. SNPs calling among the 6 isolates belonging to ST11685 was done using CSI Phylogeny (Kaas et al. 2014) by using as reference strain the first isolate (isolate 56).
Results
All 7 isolates were positive for blaKPC-2 and blaBEL-1 genes. All of them showed resistance to penicillins combined with beta-lactamase inhibitors, third-generation cephalosporins, ertapenem and nalidixic acid, being susceptible to fosfomycin, tigecycline, trimethoprim/sulfamethoxazole, amikacin, gentamycin, nitrofurantoine. All yielded positive imipenem hydrolysis and KPC positive reaction with NG Carba 5. All isolates belonged to phylogroup A and two different STs were observed: 6 isolates belonged to ST11685 (recovered from April to August 2018 and January 2019) and the other isolate belonged to ST2795 clones (recovered in January 2019). One of the ST11685 was susceptible to tobramycin, and the rest of isolates were resistant. The 6 isolates belonging to the same ST (ST11685) differed in 37–226 SNPs between them (Figure S1b). Three J53 transconjugants were obtained from clone ST11685 and one from the isolate of clone ST2795. All isolates and transconjugants were susceptible to cefiderocol (MIC range ≤ 0.03–0.25 mg/l). The transconjugants from ST11685 showed 1 dilution higher MIC value (0.125 mg/l) than J53 strain (0.06 mg/l) and the transconjugant from ST2795 yielded lower MIC value (≤ 0.03 mg/). All isolates and the three transconjugants were resistant to ceftolozane/tazobactam (MIC values > 2 mg/l).
Analyzing the hybrid genome obtained from both short-read and long-read sequencing methods, it was found that ST11685-56 harbored a circular sequence containing IncN-blaBEL-1 of 77,5 kb (p56N-BEL-1) belonging to ST24/MOBF and a circular sequence containing IncP-blaKPC-2 of 43 kb (p56P-KPC-2). The comparison by using BLASTN of IncN-blaBEL-1 and IncP-blaKPC-2 from ST11685-56 with plasmid Illumina sequences from the other ST11685 isolates yielded a homology > 99% in all cases. Additionally, in one of the ST11685 isolates was also detected an IncX5 replicon (Figure S1a). The ST2795-410 hybrid genome showed a larger IncN-blaBEL-1 of 109 kb (p410N-BEL-1), also belonging to ST24/MOBF and a IncP-blaKPC-2 (p410P-KPC-2) of similar size (39 kb) than ST11685 isolates. Additionally, ST2795-410 strain harbored two other plasmids which did not contain any bla gene: a IncF plasmid of 71 kb with formula F100:A-:B- and another plasmid of 251 kb which showed > 99% homology with 173 kb of a plasmid of pKPC-CAV1321 found in a C. freundii isolate detected in UK (Accession no. CP011611) (14).
Comparison of the two IncN-blaBEL-1 plasmids from each E. coli strain showed a shared region of 76,2 kb (99% identity) which contained the blaBEL-1 environment (Fig. 1A). According to the Unicycler results, the coverage of the p56N-BEL-1 and p410N-BEL-1 plasmids, standardized to the chromosome contig, was 2.31X and 3.61X, respectively. Additionally, the p410N-BEL-1 incorporated phage sequences (23 kb in total) which were shown a 98% of homology to P1-like bacteriophage RCS47 (accession no. NC_042128) (15). The existence of the N plasmid sequences alongside the P1-like sequences was verified by analyzing only reads > 20,000 bp (Figure S2). The p410N-BEL-1 included the characteristic repA of IncY of this phage as well as some genes of the baseplate and tail, including the lytic RepL and lysogenic proteins, but some parts of the 5 characteristic regions of this type of phages are missing. In the P-1-like part of the plasmid the region 1 had the mod gene disrupted by IS5, important region 3 structures were absent (c-segment, baseplate and tube genes, and immunity-associated genes) as well as region 5 genes related to head structure and processing. IncN-blaBEL-1 plasmids exhibited a completely different backbone than the previously described 12 kb- pKP-M1144 found in K. pneumoniae in Portugal. A comparison of both IncN plasmids with those deposited at NCBI revealed a 99.7% homology with a 48.3 kb region in a IncN plasmid (accession no. NZ_LT599827) carrying blaKPC-2 gene, found in an E. coli ST362 isolate detected in Germany in 2010. On the other hand, the comparison of IncP-blaKPC-2 plasmids from the two E. coli strains yielded 99,9% of identity between them. These plasmids were identical to pA1705-KPC (accession no. NZ_MH909348) by BLASTN, a plasmid of 42 kb detected in a K. pneumoniae in China (Fig. 1B). In addition to the blaKPC-2 gene flanked by ISKpn6 and ISKpn7, all these IncP plasmids, including the one from China, carried the blaTEM-1 and mer operon genes.
A BRIG comparison of the two IncN plasmids harbouring blaBEL-1 from E. coli strains analyzed in this study. Each plasmid is a ring and the larger p410N-BEL-1 was used as reference. The layout of blaBEL-1 is highlighted with a red arrow. Other antibiotic resistance determinant arrows are highlighted in blue, the insertion sequences are highlighted in green, P1-like phage sequences in gray and other genes are in purple. 1B) BRIG comparison of pA1705-KPC (blue ring) with the two IncP plasmids harboring blaKPC-2 from E. coli strains analyzed in this study, and pA1705-KPC (blue ring) was used as reference. The layout of blaKPC-2 highlighted with a blue arrow, the operon mer genes in gray, and the rest of genes in the same colors as in A
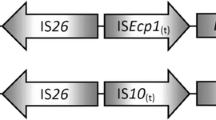
Both in ST11685-56 and ST2795-410 E. coli strains, the blaBEL-1 gene was found embedded in a class 3 integron. Surrounding the blaBEL-1 gene, the gene fosE was found upstream and blaOXA-10 and aac(6’)-1b genes were identified downstream. In addition, this class 3 integron was surrounded by Tn3 transposon sequences. This cassette was found to be homologous by BLASTN search to another structure found in a Citrobacter freundii complex isolate (121SC57), collected from a sewage sample in Spain in 2012 (Accession no. DACUGO010000000.1) (16). Within this isolate, two copies of the class 3 integron structure containing the blaBEL-1 gene, with the same gene content that in our E. coli strains, were found (Fig. 2). The chromosomal or plasmid location of these two copies in the C. freundii isolate is unknown as the contigs where they were found were too small, but the mobA and repA genes were also present in the contigs of both copies and could indicate a possible plasmid localization. The analysis of the mobA sequence assigned these two contigs to MOBQ protein family. Additionally, analyzing the plasmid content of the C. freundii genome using PlasmidFinder the following Col440I, IncFIB(K), IncN, IncP6, IncR and pKPC-CAV1321 replicons were identified.
Comparison of blaBEL-1 containing genetic environments of Enterobacterales compared in this study. Protein coding regions are represented by arrows indicating the direction of transcription and colored as follows: BEL-1 in red, other resistance determinants in blue, integrase in pink, insertion sequences and transposases in green, the relasaxe and RepA protein in yellow, and hypothetical proteins in orange. The gray shedding reflects nucleotide sequence identities. The alignments were represented with EasyFig v2.2.5
When the Portuguese K. pneumoniae isolate was compared with the E. coli BEL-1 platforms, the blaBEL-1 gene was similarly located within an identical class 3 integron, but in this case in a 12 kb ColE-1 like plasmid (pKP-M1144, accession no. KF745070), and the integron contained a blaIMP-8 and a blaGES-5 genes upstream the blaBEL-1 gene, fosE and blaOXA-10 were not found and a Tn5403 was detected downstream (Fig. 2). Finally, also using BLASTN search, an isolate of Raoultella ornithinolytica (accession no. DACSDR010000000.1) carrying BEL-1 gene was found, having this genome a short contig containing only the BEL-1 gene and a IS6100 insertion sequence.
Discussion
This is the first description of BEL-1 and KPC-2 co-produced in Enterobacterales and the first detection of this enzyme in E. coli. Although the origin of this combination is unknown, it is most probable that these strains originated from the discharge of a patient. However, both the clonal relationship of the isolates and the characteristics of the plasmids and cassettes may also indicate the possibility that that they may be the result of gene transfers and rearrangements in a human spill environment, rather than directly from patient discharges. IncN-BEL-1 plasmids showed some environmental characteristics. Firstly, the incompatibility group IncN is very abundant in wastewater of human origin, both municipal and hospital sources. In a study conducted in the La Paz River basin in Bolivia, where hospitals, industries, and households discharge directly, all the plasmids detected from the urban site were IncN (Guzman-Otazo et al. 2022). A comparative study of the inlet of a municipal wastewater treatment plant (WWTP) and the main sewer line of a Swedish hospital also revealed that IncN plasmids constituted the majority of the microbial population (Hutinel et al. 2021). This group of plasmids are self-conjugative, common in Enterobacterales and they had been harbored ESBLs, carbapenemases, and aminoglycoside-modifying enzymes (García-Fernández et al. 2011). On the other hand, IncP-KPC-2 plasmids are broad-spectrum conjugative plasmids, and have been localized in Enterobacteriaceae recovered from hospital waste in Japan (Ota et al. 2022).
The second notable feature is the type 3 integrase carrying blaBEL-1 that we found in our isolates, which is also present in the previous C. freundii and K. pneumoniae isolates. To date, the blaBEL-1 has usually been found in cassettes with class 1 integrons in P. aeruginosa (Bouheraoua et al. 2018), and had only been observed associated with the class 3 integrase in the previously reported K. pneumoniae isolate from Portugal (Papagiannitsis et al. 2015). The class 3 integrase is not frequent in collections of multidrug-resistant isolates and has not been detected in isolates from healthy volunteers in Spain (Vinué et al. 2008) or in a population-based study in France (Laroche et al. 2009). In contrast, in a study of wastewater samples from a municipal treatment plant in Canada (Jankowski et al. 2022), the class 3 integrase was found almost as frequently as class 1, which reinforces the hypothesis that the cassette with BEL-1 is found in isolates adapted to wastewater. When we compared the genetic environment of blaBEL-1 from our environmental isolates and the one from the clinical Portuguese isolate, the differences found in the two class 3 integrase cassettes may indicate different captures or two different evolutions of this cassette.
Finally, the E. coli strains are detected in discharges of a hospital where no BEL-1 or KPC-2-producing isolates were detected before or during that period neither have been submitted to the regional reference laboratory of Andalucía from other hospitals, where submissions are voluntary. It cannot be ruled out that there are colonized or infected patients and that they have not been detected, since not all E. coli isolates recovered from the hospital have been characterized at the molecular level. The persistence of one of the E. coli strains throughout the year, albeit with small differences between isolates, also supports the possibility that these isolates could be part of the collector biofilms.
Parallel to the arguments supporting a human sewage origin, a feature that may indicate a human origin of blaBEL-1 gene is the presence of P1-like phage sequences in the plasmid of one of the two strains. P1-like prophages can behave as a plasmid due to a plasmid replicon which belongs to plasmid incompatibility group (Inc) Y, and several reports have associated P1-like elements with mcr-1 and ESBL genes (Billard-Pomares et al. 2014). What has been described to date are isolates that carry P1-like phage-plasmids in addition to other plasmids. Fusions of plasmids and P1-like phage-plasmids have only previously been demonstrated in an E. coli isolate obtained from a fecal sample of an inpatient in China (Bai et al. 2017). On this occasion, it was found the fusion of IncH12 with P1 and, similar to our strain, the phage sequences were incomplete.
Finally, it remains to comment that these isolates are susceptible to cefiderocol despite the production of BEL-1. The presence of this enzyme had been associated with a 1 dilution-increase in the MIC value of cefiderocol when this gene was cloned into pUCp24 (Poirel et al. 2022). A similar increase was observed in some of the transconjugants of clone ST11685, but this increase does not seem to confer resistance.
Conclusions
In conclusion, we have found the presence of BEL-1 producing E. coli isolates associated with KPC-2 production in hospital discharges with some genetic characteristics typical of human wastewater resident flora and in a different cassette than usually observed in P. aeruginosa. Broader epidemiological studies, including environmental samples from other non-human sources, would be necessary to determine the origin of this new BEL-1 cassette.
Data availability
All datasheets of this study will be available at https://idus.us.es after publication.
References
Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA (2011) BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. https://doi.org/10.1186/1471-2164-12-402
Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS (2016) PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. https://doi.org/10.1093/nar/gkw387
Bai L, Wang J, Hurley D, Yu Z, Wang L, Chen Q, Li J, Li F, Fanning S (2017) A novel disrupted mcr-1 gene and a lysogenized phage P1-like sequence detected from a large conjugative plasmid, cultured from a human atypical enteropathogenic Escherichia coli (aEPEC) recovered in China. J Antimicrob Chemother 72:1531–1533. https://doi.org/10.1093/jac/dkw564
Billard-Pomares T, Fouteau S, Jacquet ME, Roche D, Barbe V, Castellanos M, Bouet JY, Cruveiller S, Médigue C, Blanco J, Clermont O, Denamur E, Branger C (2014) Characterization of a P1-like bacteriophage carrying an SHV-2 extended-spectrum β-lactamase from an Escherichia coli strain. Antimicrob Agents Chemother 58:6550–6557. https://doi.org/10.1128/AAC.03183-14
Bouheraoua N, Poirel L, Bourreau B, Bonnin R, Laroche L, Naas TNP (2018) Integrase-mediated recombination of the bel-1 gene cassette encoding the extended-spectrum β-lactamase BEL-1. Antimicrob Agents Chemother 62:e00030-e118. https://doi.org/10.1128/AAC.00030-18
Brolund A, Sandegren L (2016) Characterization of ESBL disseminating plasmids. Infect Dis (auckl) 48:18–25. https://doi.org/10.3109/23744235.2015.1062536
Doi Y (2019) Treatment options for carbapenem-resistant Gram-negative bacterial infections. Clin Infect Dis 69:S565–S575. https://doi.org/10.1093/cid/ciz830
Falcone M, Tiseo G (2022) Cefiderocol for the treatment of metallo-β-Lactamases producing Gram-negative bacilli: lights and shadows from the literature. Clin Infect Dis 75:1085–1087. https://doi.org/10.1093/cid/ciac082
Falgenhauer L, Ghosh H, Doijad S, Yao Y, Bunk B, Spröer C, Kaase M, Hilker R, Overmann J, Imirzalioglu C, Chakraborty, (2017) Genome analysis of the carbapenem- and colistin-resistant Escherichia coli isolate NRZ14408 reveals horizontal gene transfer pathways towards panresistance and enhanced virulence. Antimicrob Agents Chemother 61:e02359-e2416. https://doi.org/10.1128/AAC.02359-16
García-Fernández A, Villa L, Moodley A, Hasman H, Miriagou V, Guardabassi L, Carattoli A (2011) Multilocus sequence typing of IncN plasmids. J Antimicrob Chemother 66:1987–1991. https://doi.org/10.1093/jac/dkr225
Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI (2014) Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. MBio 5:e01918–4. https://doi.org/10.1128/mBio.01918-14
Guzman-Otazo J, Joffré E, Agramont J, Mamani N, Jutkina J, Boulund F, Hu YOO, Jumilla-Lorenz D, Farewell A, Larsson DGJ, Flach CF, Iñiguez V, Sjöling Å (2022) Conjugative transfer of multi-drug resistance IncN plasmids from environmental waterborne bacteria to Escherichia coli. Front Microbiol 13:1–18. https://doi.org/10.3389/fmicb.2022.997849
Hubeny J, Harnisz M, Korzeniewska E, Buta M, Zieliński W, Rolbiecki D, Giebułtowicz J, Nałęcz-Jawecki G, Płaza G (2021) Industrialization as a source of heavy metals and antibiotics which can enhance the antibiotic resistance in wastewater, sewage sludge and river water. PLoS ONE 16:1–24. https://doi.org/10.1371/journal.pone.0252691
Hutinel M, Fick J, Larsson DGJ, Flach CF (2021) Investigating the effects of municipal and hospital wastewaters on horizontal gene transfer. Environ Pollut 276:116733. https://doi.org/10.1016/j.envpol.2021.116733
Jankowski P, Gan J, Le T, McKennitt M, Garcia A, Yanaç K, Yuan Q, Uyaguari-Diaz M (2022) Metagenomic community composition and resistome analysis in a full-scale cold climate wastewater treatment plant. Environ Microbiomes 17:1–20. https://doi.org/10.1186/s40793-022-00398-1
Juan C, Zamorano L, Pérez JL, Ge Y, Oliver A (2010) Activity of a new antipseudomonal cephalosporin, CXA-101 (FR264205), against carbapenem-resistant and multidrug-resistant Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 54:846–851. https://doi.org/10.1128/AAC.00834-09
Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O (2014) Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 9:1–8. https://doi.org/10.1371/journal.pone.0104984
Korzeniewska E, Harnisz M (2013) Beta-lactamase-producing Enterobacteriaceae in hospital effluents. J Environ Manage 123:1–7. https://doi.org/10.1016/j.jenvman.2013.03.024
Laroche E, Pawlak B, Berthe T, Skurnik D, Petit F (2009) Occurrence of antibiotic resistance and class 1, 2 and 3 integrons in Escherichia coli isolated from a densely populated estuary (Seine, France). FEMS Microbiol Ecol 68:118–130. https://doi.org/10.1111/j.1574-6941.2009.00655.x
Liu C, Li B, Wu BB, Lin H, Jiang L, Qiu Y (2022) How heavy metal stress promotes dissemination of antibiotic resistance genes in the activated sludge process. J Hazard Mater 437:129279. https://doi.org/10.1016/j.jhazmat.2022.129279
Ota Y, Prah I, Nukui Y, Koike R, Saitoa R (2022) bla KPC-2 -Encoding IncP-6 Plasmids in Citrobacter freundii and Klebsiella variicola strains from hospital sewage in Japan. Appl Environ Microbiol 88:e00019. https://doi.org/10.1128/aem.00019-22
Papagiannitsis CC, Dolejska M, Izdebski R, Dobiasova H, Studentova V, Esteves FJ, Derde LPG, Bonten MJM, Hrabák J, Gniadkowski M (2015) Characterization of pKP-M1144, a novel ColE1-like plasmid encoding IMP-8, GES-5, and BEL-1 β-lactamases, from a Klebsiella pneumoniae sequence type 252 isolate. Antimicrob Agents Chemother 59:5065–5068. https://doi.org/10.1128/AAC.00937-15
Poirel L, Brinas L, Verlinde A, Ide L, Nordmann P (2005) BEL-1, a novel clavulanic acid-inhibited extended-spectrum β-lactamase, and the class 1 integron In120 in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:3743–3748. https://doi.org/10.1128/AAC.49.9.3743-3748.2005
Poirel L, Docquier JD, De Luca F, Verlinde A, Ide L, Rossolini GM, Nordmann P (2010) BEL-2, an extended-spectrum β-lactamase with increased activity toward expanded-spectrum cephalosporins in Pseudomonas aeruginosa. Antimicrob Agents Chemother 54:533–535. https://doi.org/10.1128/AAC.00859-09
Poirel L, De la Rosa JMO, Sadek M, Nordmann P (2022) Impact of acquired broad-spectrum β-lactamases on susceptibility to cefiderocol and newly developed β-lactam/β-lactamase inhibitor combinations in Escherichia coli and Pseudomonas aeruginosa. Antimicrob Agents Chemother 66:e0003922. https://doi.org/10.1128/AAC.00039-22
Robertson J, Nash JHE (2018) MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genomics 4:e000206. https://doi.org/10.1099/mgen.0.000206
Romero-Oraá L, Borrego J, Galán F, Tejero R, Rojo MD, Pascual A, López-Cerero L (2019) Multi-centre monitoring of acquired carbapenemases-producing Gramnegative bacteria from hospital sewages in the South of Spain: results from the Canalis Project. Seville, Spain. 29th European congress of clinical microbiology and infectious diseases, Amsterdam, Netherlands. abst O0053:31
Vinué L, Sáenz Y, Somalo S, Escudero E, Moreno MÁ, Ruiz-Larrea F, Torres C (2008) Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J Antimicrob Chemother 62:934–937. https://doi.org/10.1093/jac/dkn331
Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:1–22. https://doi.org/10.1371/journal.pcbi.1005595
Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS (2011) PHAST: a fast phage search tool. Nucleic Acids Res 39:347–352. https://doi.org/10.1093/nar/gkr485
Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M (2020) The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res 30:138–152. https://doi.org/10.1101/gr.251678.119
Funding
Funding for open access publishing: Universidad de Sevilla/CBUA. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. This work was supported by the Plan Nacional de I1D1i 2013–016 and the Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad (PI17/02003). M.R.P. is supported by a VIPPI-US fellowship from the University of Seville.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection was performed by Laura Romero-Oraá, Fatima Galán, and María Victoria García Palacios. Analysis of plasmid and figures was carried out by Lorena López-Cerero and Marina R. Pulido. The first draft of the manuscript was written by Lorena López-Cerero and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Diane Purchase
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Romero-Oraá, L., Pulido, M.R., Galán, F. et al. Genetic features of BEL-1-producing and KPC-2-producing E. coli from hospital wastewater: human source or sewages adaptation. Environ Sci Pollut Res 31, 43896–43902 (2024). https://doi.org/10.1007/s11356-024-33875-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-024-33875-w