Abstract
Legionella pneumophila (L. pneumophila) is one of the main pathogens, causing pneumonia and respiratory tract infections, especially in patients with ventilator-associated pneumonia (VAP). This study aimed to approve the hypothesis that the serogroup distribution of L. pneumophila isolates from patients is correlated with Legionella strains in the environment. A total of 280 bronchoalveolar lavage (BAL) samples from VAP patients admitted to the intensive care unit (ICU) as well as 116 water samples from different sources in four hospitals in Tehran, Iran, were evaluated for the presence of L. pneumophila infection by culture, nested polymerase chain reaction (PCR), real-time PCR, and sequencing for genetic diversity. The molecular and culture methods found 24 (8.6%) and 5 (1.8%) samples to be positive for L. pneumophila in VAP patients, while they found 23 (19.8%) and 8 (6.9%) positive samples in water resources, respectively. The sequencing results indicated that all positive clinical samples and 14 (60.8%) environmental samples were belonged to L. pneumophila serogroup 1. Smoking, age, length of ICU stay, and duration of ventilator use had strong relationship with L. pneumophila infectivity. In conclusion, this is the first report from Iran to determine minor differences in the serogroup distribution of environmental and clinical strains. However, further studies are needed to confirm this relationship in different regions of Iran.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ventilator-associated pneumonia (VAP) is the most common intensive care unit (ICU)-acquired infection and a leading cause of mortality in patients admitted to ICUs (Al-Omari et al. 2021).
Bacterial agents related to VAP vary depending on several factors, such as duration of mechanical ventilation, local ecology, timing and cumulative exposure to antimicrobials, hospitalization, ICU stays before VAPs, and occurrence of any potential epidemic phenomena in the ICU setting. Common gram-negative bacteria associated with VAP include Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter species, and Escherichia coli; Staphylococcus aureus, and the main gram-positive bacterium are Staphylococcus aureus. Other bacteria, such as Mycoplasma pneumoniae and Legionella pneumophila (L. pneumophila), have been also detected (Papazian et al. 2020; Torres et al. 2021).
Several studies have indicated that 2–15% and above 30% of community- and hospital-acquired pneumonia infections are caused by L. pneumophila, respectively (Chahin & Opal 2017; Torres et al. 2021). However, L. pneumophila cannot be distinguished from other pneumonia infections according to the clinical signs. Overall, early diagnosis and treatment of infection can be helpful in reducing the mortality rate (Khosroshahi et al. 2015).
The severity of legionellosis varies from a mild febrile disease to a severe and even fatal form of pneumonia, caused by exposure to L. pneumophila in water. The most common route of L. pneumophila transmission is inhalation of contaminated aerosols. Aerosol sources correlated with Legionella transmission include hot- and cold-water systems, humidifiers, whirlpool spas, and air conditioning cooling towers. These sources are known to involve the respiratory system. Individuals, especially susceptible hospitalized patients, can be also infected with contaminated water or aspiration of ice water (Gonçalves et al. 2021; Torres et al. 2021).
Although the incidence of legionellosis has been reported worldwide, its exact occurrence remains undetermined due to inappropriate monitoring of infected cases. Early diagnosis of this disease depends on the physician’s increased awareness of the disease and availability of the required equipment for detection (Yaslianifard et al. 2019).
The use of molecular epidemiological techniques to characterize clinical and environmental isolates can provide data for specific geographical areas and allow phylogenetic and population analyses. Overall, identification of these characteristics is essential for a better understanding of the transmission routes of bacteria in the environment. The sequence type distribution of L. pneumophila in North America, Europe, and Asia revealed the predominance of specific sequence types in the clinical isolates and indicated that most clinical cases and outbreaks were caused by a decrease in the number of L. pneumophila isolates (Bianchi et al. 2016; Quero et al. 2018).
With this background in mind, the current study aimed to evaluate and compare clinical and environmental L. pneumophila isolates from water resources in four hospitals of Tehran, Iran.
Methods
Sample collection from VAP patients and culture
In this study, 280 bronchoalveolar lavage (BAL) samples were collected from intubated patients, admitted to the ICUs of four teaching hospitals affiliated to Tehran University of Medical Sciences, Tehran, Iran. The fresh samples were divided into two parts; one part was used for cultivation, and the other part was used for the polymerase chain reaction (PCR) assay (stored at -20 °C until testing).
Before culture, sample decontamination was required to decrease the contaminating microbiota in respiratory samples by diluting (1:10) the BAL samples in a low-pH KCl/HCl buffer (pH 2.2) and incubating them at room temperature for four minutes before plating the suspension onto a buffered charcoal yeast extract (BCYE) agar medium (Sigma Aldrich, France). The plates were incubated at 37 °C in a 5% CO2 atmosphere with humidity and kept for one week (Edelstein and Lück 2015). Also, L. pneumophila ATCC33152 was used as a positive control. Bacterial colonies emerged following the fifth day.
Sample collection from water resources and cultures
In the present study, 116 water samples from ICUs, where VAP patients were hospitalized; were collected from different water sources, such as water tap, air conditioners, and drinking water coolers in sterile, non-toxic 1000-mL plastic bottles; and then transferred to the laboratory in less than 4 h.
The water samples were centrifuged at 3000 g for 30 min (25 ℃). Next, 500 µL of precipitation was treated with 4.5 mL of acid wash buffer, as mentioned above. After shaking, 200 μL of suspension was inoculated on the BCYE agar, supplemented with glycine, vancomycin, cyclohexamide, and polymyxin B (Edelstein and Lück 2015).
The drug susceptibility test (DST) for antimicrobial agents was also performed using the disk diffusion method for all positive culture isolates from clinical and environmental samples (Fard et al. 2012, Rahimi and Vesal 2017).
Nested PCR and real-time PCR methods
Purified genomic DNA was extracted from BAL samples, water samples, and bacterial colonies using a PROBA-NK DNA Extraction Kit DNA-Technology Company, Moscow, Russia), according to the manufacturer’s instructions.
In this study, we identified Legionella species by both nested and real-time PCR assays, using the macrophage infectivity potentiator (mip) gene primer, as previously described (Fiume et al. 2005; Wilson et al. 2003). The selection of mip gene for screening was based on its discriminative power and frequent use in other studies (Ahmadrajabi et al. 2016).
DNA sequencing and phylogenetic analysis
For L. pneumophila genotyping, the mip gene from the isolates was amplified using primers, Legmip_f and Legmip_r, as previously described (Ratcliff et al. 1998). First, a High-Pure PCR Product Purification Kit (Sigma-Aldrich, Missouri, USA) was used for the purification of PCR products. Next, an automated ABI sequencer (Applied Biosystems, Foster City, CA, USA) was applied for PCR sequencing. Genotyping was performed using a phylogenetic analysis, with reference sequences selected from the GenBank database. Moreover, a phylogenetic tree reconstruction was performed in MEGA version 6.0 software (http://www.megasoftware.net), using the neighbor joining method and K2P model with 500 bootstrap replicates.
Statistical analysis
SPSS for Windows Version 22.0 (2016; IBM Corp., Armonk, NY, USA) was applied for data analysis. The normal distribution of continuous variables was determined by Shapiro–Wilk test. Pearson’s Chi-square and Mann–Whitney U tests were also performed to evaluate quantitative and continuous variables, respectively. Besides, for evaluating the relationships between the risk factors and L. pneumophila infectivity, a multivariate logistic regression analysis was carried out using the Hosmer–Lemeshow test. Two-tailed P value > 0.05 was considered statistically significant.
Results
Detection of L. pneumophila in VAP patients
Of 280 BAL samples, 24 (8.6%) were positive in nested and real-time PCR. Of 24 PCR-positive samples, five (20.8%) showed growth in the BCYE agar. These patients (A1, A2, B1, C1, and D1) were infected with a large number of bacteria in the culture. The direct fluorescent antibody (DFA) test revealed that these isolates belonged to the L. pneumophila serogroup 1.
The mean age of positive cases was 69.5 ± 20.3 years, and the male-to-female ratio was 2.0 (16 vs. 8). The mean duration of ventilator use and ICU stay were 26.5 ± 7.6 and 33.0 ± 7.8 days, respectively (Table 1). Also, among 24 positive cases, 18 (75%) were smokers.
All positive samples were examined to determine the most common viral, fungal, and bacterial infectious agents of the respiratory tract, such as Mycobacterium tuberculosis, Pseudomonas aeruginosa, Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, Candida albicans, Aspergillus spp., Mucor spp., influenza A and B viruses, coronaviruses, and rhinoviruses. All positive samples were negative for these agents.
The DST revealed that all five isolates were susceptible to rifampicin (10 μg), levofloxacin (5 μg), tetracycline (15 μg), clarithromycin (15 μg), and erythromycin (15 μg) and resistance to streptomycin (10 μg), kanamycin (30 μg), and doxycycline (30 μg).
Detection of L. pneumophila in water resources
Out of 116 water samples, 23 (19.8%) and 8 (6.9%) were positive for L. pneumophila based on PCR and culture methods, respectively; this indicates the higher sensitivity of molecular methods compared to the culture method. The L. pneumophila distribution in water resources is shown in Table 2. In this study, the most common contaminated sources in the hospitals included water in ICUs and tap water. All L. pneumophila isolates from cultures belonged to serogroup 1, as determined by the DFA test.
The DST revealed that all eight isolates were susceptible to rifampicin (10 μg), levofloxacin (5 μg), and erythromycin (15 μg).
Phylogenetic analysis of L. pneumophila by genotyping the mip gene
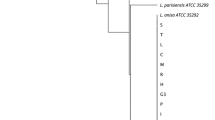
To confirm genotyping by the DFA test, the PCR product of the mip gene was sequenced as previously described (Ratcliff et al. 1998). All positive samples based on nested and real-time PCR were amplified and sequenced to construct a phylogenetic tree. The sequencing results of the clinical samples revealed that all positive samples belonged to serogroup 1, while in environmental samples, only 14 (60.8%) belonged to serogroup 1; other samples belonged to serogroups 2, 3, 8, and 10. The sequences of L. pneumophila indicated more than 98% homology to serogroup 1 sequences available in the GenBank database (Fig. 1).
Factors related to L. pneumophila infectivity
We performed a multivariate logistic regression to assess the correlations between the clinical parameters and L. pneumophila infectivity. The factors independently related to L. pneumophila infectivity include age (OR, 1.164; 95% CI, 1.009–3.089; P = 0.010), smoking (OR, 1.895; 95% CI, 1.001–1.995; P = 0.025), length of ICU stay (OR, 0.951; 95% CI, 0.885–0.998; P < 0.001), and duration of ventilator use (OR, 0.921; 95% CI, 0.863–0.984; P < 0.001) (Table 3).
Discussion
In this preliminary study, we investigated and compared the clinical and environmental L. pneumophila serogroup isolates in four hospitals. According to our findings, the serogroup distribution of L. pneumophila isolates from patients was almost similar to the distribution of L. pneumophila isolates from the environment. Previous reports from different regions have suggested a possible similarity between these distribution patterns (Kozak-Muiznieks et al. 2014; Quero et al. 2018), although they have not been performed based on a systematic collection of both environmental and patient Legionella isolates.
To the best of our knowledge, this is the first study from Iran to evaluate the relationship between the serogroup diversity of L. pneumophila isolates collected from patients and environmental samples.
L. pneumophila was identified in 8.4% and 19.8% of patients and water resources in our study, respectively. Generally, water supply systems are an important source of infection. According to several reports from Iran, the prevalence of Legionella species in water resources is high, and L. pneumophila is the most prevalent species (Khaledi et al. 2019; Yaslianifard et al. 2019). The prevalence of this pathogen is estimated at 9.6% in Iranian clinical samples, ranging from 0.4 to 22.1% (Khaledi et al. 2019). According to previous results, this agent could be important for Iranian patients with respiratory problems. Therefore, use of an efficient diagnostic method can help with a timely treatment of patients’ symptoms (Goodarzi et al. 2020).
In the present study, L. pneumophila serogroup 1 was responsible for VAP infection. In this regard, several studies have shown that most patients with Legionnaires’ disease (LD) were infected with serogroup 1 isolates. These results suggest that serogroup 1 is more virulent than other serogroups (Den Boer et al. 2008). Also, 60.8% of positive samples from water resources belonged to serogroup 1 in our study. Based on other reports from different regions of Iran, serogroup 1 is more frequently found in water resources (Moosavian et al. 2019; Yaslianifard et al. 2019), although it varies in other countries (Den Boer et al. 2008; Harrison et al. 2007). The distribution of legionella isolates may depend on the location and geographical area, as shown in a study from Korea (Lee et al. 2010).
Unlike other studies, environmental isolates in our study showed similar Legionella genotypes to those collected from the patients. There are two possible explanations for this finding. First, surveys based on standardized questionnaires may not be able to determine the actual source of LD. It is also reasonable to assume that the collection of isolates from the environment represents the country. However, it is feasible that virulent strains should be examined in the air, but not in aquatic sources, as they spread through different sources on days of increased humidity (Fisman et al. 2005). Second, the genotypes of Legionella, most commonly isolated from human respiratory secretions, are also present in sampling sources, although their concentration cannot be determined (Den Boer et al. 2008).
In the present study, age, length of ICU stay, duration of ventilator use, and smoking were significantly associated with L. pneumophila infectivity. Several studies have shown that the infectivity of L. pneumophila is related to the host and different strains. For example, the demographic and clinical characteristics of patients infected with two common isolates (Paris and Lorraine) were different in France. The Paris strain was significantly related to a history of cancer or hematologic malignancy, steroid therapy, and female sex, while it was negatively correlated with smoking. On the other hand, the Lorraine strain was only related to smoking. The crude mortality rate correlated with the Paris strain was higher than that of the sporadic strain, while the Lorraine strain was associated with a lower mortality rate than the sporadic strain (Ananth and Kleinbaum 1997; Ginevra et al. 2009).
The known host risk factors associated with LD include an immunocompromised status, diabetes, older age, being a recipient of a transplant or chemotherapy, smoking, and chronic obstructive pulmonary disease (Farnham et al. 2014). Also, the most important environmental risk factors include proximity to cooling towers, decorative fountains, and whirlpool spas, residence in a healthcare facility, and traveling (Beauté et al. 2013; Greig et al. 2004).
Conclusion
This is the first report from Iran to evaluate differences in the serogroup distribution of environmental and clinical strains of L. pneumophila. We found that serogroup 1 was the most common type in the samples. However, further studies in different regions of Iran are warranted to determine the relationship between environmental and clinical strains.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- L. pneumophila:
-
Legionella pneumophila
- VAP:
-
Ventilator-associated pneumonia
- BAL:
-
Bronchoalveolar lavage
- ICU:
-
Intensive care unit
- PCR:
-
Polymerase chain reaction
- BCYE:
-
Buffered charcoal yeast extract
- DST:
-
Drug susceptibility test
- Mip :
-
MAcrophage infectivity potentiator
- DFA:
-
Direct fluorescent antibody
References
Ahmadrajabi R, Shakibaie MR, Iranmanesh Z, Mollaei HR, Sobhanipoor MH (2016) Prevalence of mip virulence gene and PCR-base sequence typing of Legionella pneumophila from cooling water systems of two cities in Iran. Virulence 7:602–609
Al-Omari B, McMeekin P, Allen AJ, Akram AR, Graziadio S, Suklan J, Jones WS, Lendrem BC, Winter A, Cullinan M (2021) Systematic review of studies investigating ventilator associated pneumonia diagnostics in intensive care. BMC Pulm Med 21:1–19
Ananth CV, Kleinbaum DG (1997) Regression models for ordinal responses: a review of methods and applications. Int J Epidemiol 26:1323–1333
Beauté J, Zucs P, De Jong B (2013) Legionnaires’ disease in Europe, 2009–2010. Eurosurveillance 18:20417
A Bianchi, FE Pregliasco, M Consonni, M Tesauro (2016): Genotypic diversity of Legionella pneumophila in environmental and clinical strains assessed by Sequence-Based Typing, in association with retrospective clinical surveillance in Northern Italy. Annals of Agricultural and Environmental Medicine 23
Chahin A, Opal SM (2017) Severe pneumonia caused by Legionella pneumophila: differential diagnosis and therapeutic considerations. Infect Dis Clin 31:111–121
Den Boer JW, Bruin JP, Verhoef LP, Van der Zwaluw K, Jansen R, Yzerman E (2008) Genotypic comparison of clinical Legionella isolates and patient-related environmental isolates in The Netherlands, 2002–2006. Clin Microbiol Infect 14:459–466
Edelstein PH, Lück C (2015) Legionella, Manual of Clinical Microbiology, 11th edn. Wiley, New York, pp 887–904
Fard S, Nomanpour B, Fatolahzadeh B, Mobarez A, Darban-Sarokhalil D, Fooladi A, Leeuwen W, Feizabadi M (2012) Hospital acquired pneumonia: comparison of culture and real-time PCR assays for detection of Legionella pneumophila from respiratory specimens at Tehran hospitals. Acta Microbiol Immunol Hung 59:355–365
Farnham A, Alleyne L, Cimini D, Balter S (2014) Legionnaires’ disease incidence and risk factors, New York, New York, USA, 2002–2011. Emerg Infect Dis 20:1795
Fisman DN, Lim S, Wellenius GA, Johnson C, Britz P, Gaskins M, Maher J, Mittleman MA, Victor Spain C, Haas CN (2005) It’s not the heat, it’s the humidity: wet weather increases legionellosis risk in the greater Philadelphia metropolitan area. J Infect Dis 192:2066–2073
Fiume L, Bucca Sabattini M, Poda G (2005) Detection of Legionella pneumophila in water samples by species-specific real-time and nested PCR assays. Lett Appl Microbiol 41:470–475
Ginevra C, Duclos A, Vanhems P, Campèse C, Forey F, Lina G, Che D, Jerome E, Che D (2009) Host-related risk factors and clinical features of community-acquired legionnaires disease due to the Paris and Lorraine endemic strains. Clin Infect Dis 49:184–191
Gonçalves IG, Simões LC, Simões M (2021) Legionella pneumophila. Trends Microbiol 29:860–861
Goodarzi NN, Pourmand M, Rajabpour M, Arfaatabar M, Mosadegh M, Mohamad SS (2020) Frequency of Mycoplasma pneumoniae, Legionella pneumophila and Chlamydia spp. among patients with atypical pneumonia in Tehran. New microbes new infect 37:100744
Greig JE, Carnie JA, Tallis GF, Zwolak B, Hart WG, Guest CS, Ryan NJ, Leydon JA, Tan AG, Gordon IR (2004) An outbreak of Legionnaires’ disease at the Melbourne Aquarium, April 2000: investigation and case–control studies. Med J Aust 180:566–572
Harrison T, Doshi N, Fry N, Joseph C (2007) Comparison of clinical and environmental isolates of Legionella pneumophila obtained in the UK over 19 years. Clin Microbiol Infect 13:78–85
Khaledi A, Esmaeili S-A, Vazini H, Karami P, Bahrami A, Sahebkar A (2019) Evaluation of the prevalence of Legionella pneumophila in Iranian clinical samples: A systematic review and meta-analysis. Microb Pathog 129:93–98
Khosroshahi ND, Farivar TN, Johari P (2015) Identification of Legionella pneumophila in intubated patients with TaqMan real time PCR. Jundishapur J Microbiol 8:e15094
Kozak-Muiznieks NA, Lucas CE, Brown E, Pondo T, Taylor TH Jr, Frace M, Miskowski D, Winchell JM (2014) Prevalence of sequence types among clinical and environmental isolates of Legionella pneumophila serogroup 1 in the United States from 1982 to 2012. J Clin Microbiol 52:201–211
Lee HK, Shim JI, Kim HE, Yu JY, Kang YH (2010) Distribution of Legionella species from environmental water sources of public facilities and genetic diversity of L. pneumophila serogroup 1 in South Korea. Appl Environ Microbiol 76:6547–6554
Moosavian M, Moradzadeh M, Ghadiri A, Saki M (2019) Isolation and Identification of Legionella spp. in environmental water sources based on macrophage infectivity potentiator (mip) gene sequencing in southwest Iran. AIMS Microbiology 5:223–231
Papazian L, Klompas M, Luyt C-E (2020) Ventilator-associated pneumonia in adults: a narrative review. Intensiv Care Med 46:888–906
Quero S, Párraga-Niño N, Barrabeig I, Sala MR, Pedro-Botet ML, Monsó E, Jané M, Sabrià M, Garcia-Núñez M (2018) Population structure of Environmental and Clinical Legionella pneumophila isolates in Catalonia. Sci Rep 8:1–9
Rahimi B, Vesal A (2017) Antimicrobial resistance properties of Legionella Pneumophila isolated from the cases of lower respiratory tract infections. Biomed Pharmacol J 10:59–65
Ratcliff RM, Lanser JA, Manning PA, Heuzenroeder MW (1998) Sequence-based classification scheme for the genus Legionella targeting the mip gene. J Clin Microbiol 36:1560–1567
Torres A, Cilloniz C, Niederman MS, Menéndez R, Chalmers JD, Wunderink RG, van der Poll T (2021) Pneumonia Na Rev Dis Prim 7:25
Wilson DA, Yen-Lieberman B, Reischl U, Gordon SM, Procop GW (2003) Detection of Legionella pneumophila by real-time PCR for the mip gene. J Clin Microbiol 41:3327–3330
Yaslianifard S, Mohammadzadeh M, Pourmand MR, Yaslianifard S, Sepehr MN, Arfaatabar M, Norouzi M (2019) Prevalence and molecular distribution of Legionella pneumophila in cold water taps across Alborz province. Iran Clin Epidemiol Glob Health 7:525–529
Acknowledgements
We would like to thank all of the patients who participated in the study.
Author information
Authors and Affiliations
Contributions
F.S and SH.M: performed the experiments and manuscript preparation. M.Z and M.H: clinical sample and data acquisition. S.D.S and F.V: analyzed data and interpreted data. A.F: designed and supervised clinical study, interpreted data, and read and approved manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was carried out at Pasteur Institute of Iran (PII; Tehran, Iran) from May 2017 to March 2018. It was performed according to the Declaration of Helsinki (1975) guidelines and local regulations and also approved by the Ethics Committee of PII. Written informed consent was directly obtained from all of the participants.
Consent for publication
Not applicable.
Competing interests
The authors declare competing interests.
Additional information
Responsible Editor: Lotfi Aleya
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sakhaee, F., Mafi, S., Zargar, M. et al. Correlation between Legionella pneumophila serogroups isolated from patients with ventilator-associated pneumonia and water resources: a study of four hospitals in Tehran, Iran. Environ Sci Pollut Res 29, 41368–41374 (2022). https://doi.org/10.1007/s11356-022-18867-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-022-18867-y