Abstract
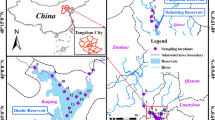
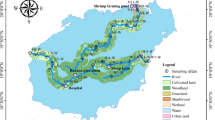
Antibiotic resistance genes (ARGs) have become an important public health problem. In this study, we used metagenomic sequencing to analyze the composition of ARGs in selected original habitats of northeast China, comprising three different rivers and riverbank soils of the Heilongjiang River, Tumen River, and Yalu River. Twenty types of ARG were detected in the water samples. The major ARGs were multidrug resistance genes, at approximately 0.5 copies/16S rRNA, accounting for 57.5% of the total ARG abundance. The abundance of multidrug, bacitracin, beta-lactam, macrolide-lincosamide-streptogramin, sulfonamide, fosmidomycin, and polymyxin resistance genes covered 96.9% of the total ARG abundance. No significant ecological boundary of ARG diversity was observed. The compositions of the resistance genes in the three rivers were very similar to each other, and 92.1% of ARG subtypes were shared by all water samples. Except for vancomycin resistance genes, almost all ARGs in riverbank soils were detected in the river water. About 31.05% ARGs were carried by Pseudomonas. Opportunistic pathogenic bacteria carrying resistance genes were mainly related to diarrhea and respiratory infections. Multidrug and beta-lactam resistance genes correlated positively with mobile genetic elements (MGEs), indicating a potential risk of diffusion. The composition of ARGs in three different rivers was similar, indicating that climate plays an important role in ARG occurrence. ARG subtypes in river water were almost completely the same as those in riverbank soil. ARGs had no significant geographical distribution characteristics. Many ARGs were carried by human pathogenic bacteria related to diarrhea and respiratory infections, such as Pseudomonas aeruginosa and Aeromonas caviae. In general, our results provide a valuable dataset of river water ARG distribution in northeast China. The related ecological and geographical distribution characteristics should be further explored.







Similar content being viewed by others
Data Availability
Not applicable.
References
Aminov RI (2011) Horizontal gene exchange in environmental microbiota. Front Microbiol 2:158
Angermeyer A, Crosby SC, Huber JA (2016) Decoupled distance-decay patterns between dsrA and 16S rRNA genes among salt marsh sulfate-reducing bacteria. Environ Microbiol 18(1):75–86
Bai Y, Ruan X, Xie X, Yan Z (2019) Antibiotic resistome profile based on metagenomics in raw surface drinking water source and the influence of environmental factor: A case study in Huaihe River Basin, China. Environ Pollut 248:438–447
Baltz RH (2008) Renaissance in antibacterial discovery from actinomycetes. Current Opinion in Pharmacology 8(5):0–563
Bao, P., Xiao, K.Q., Li, B., Ma, L., Zhou, X., Zhang, T. and Zhu, Y.G. (2016) Metagenomic profiles of antibiotic resistance genes in paddy soils from South China. FEMS Microbiol Ecol 92(3).
Bastian, M., Heymann, S. and Jacomy, M. (2009) Gephi: An Open Source Software for Exploring and Manipulating Networks.
Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, Bürgmann H, Sørum H, Norström M, Pons MN, Kreuzinger N, Huovinen P, Stefani S, Schwartz T, Kisand V, Baquero F, Martinez JL (2015) Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol 13(5):310–317
Blaak H, de Kruijf P, Hamidjaja RA, van Hoek AH, de Roda Husman AM, Schets FM (2014) Prevalence and characteristics of ESBL-producing E. coli in Dutch recreational waters influenced by wastewater treatment plants. Vet Microbiol 171(3–4):448–459
Chen B, Liang X, Nie X, Huang X, Zou S, Li X (2015) The role of class I integrons in the dissemination of sulfonamide resistance genes in the Pearl River and Pearl River Estuary, South China. J Hazard Mater 282:61–67
Chen H, Bai X, Li Y, Jing L, Chen R, Teng Y (2019) Source identification of antibiotic resistance genes in a peri-urban river using novel crAssphage marker genes and metagenomic signatures. Water Research 167:115098
Cheng, J., Tang, X. and Liu, C. (2020) Occurrence and distribution of antibiotic resistance genes in various rural environmental media. Environmental Science & Pollution Research International.
Cui P, Bai Y, Li X, Peng Z, Chen D, Wu Z, Zhang P, Tan Z, Huang K, Chen Z, Liao H, Zhou S (2020) Enhanced removal of antibiotic resistance genes and mobile genetic elements during sewage sludge composting covered with a semi-permeable membrane. Journal of Hazardous materials 396:122738
Dias MF, da Rocha Fernandes G, Cristina de Paiva M, de Matos Christina, Salim A, Santos A.B. Amaral, Nascimento AM (2020) Exploring the resistome, virulome and microbiome of drinking water in environmental and clinical settings. Water Research 174:115630
Du D, Wang-Kan X, Neuberger A, van Veen HW, Pos KM, Piddock LJV, Luisi BF (2018) Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol 16(9):523–539
He P, Wu Y, Huang W, Wu X, Lv J, Liu P, Bu L, Bai Z, Chen S, Feng W, Yang Z (2020) Characteristics of and variation in airborne ARGs among urban hospitals and adjacent urban and suburban communities: A metagenomic approach. Environ Int 139:105625
Hiroshi N, Jean-Marie P (2012) Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev 36(2):340–363
Leonard, A.F.C., Zhang, L., Balfour, A., Garside, R. and Gaze, W.H. (2015) Human recreational exposure to antibiotic resistant bacteria in coastal bathing waters. Environment International 82(sep.), 92–100.
Li A, Chen L, Zhang Y, Tao Y, Xie H, Li S, Sun W, Pan J, He Z, Mai C (2018) Occurrence and distribution of antibiotic resistance genes in the sediments of drinking water sources, urban rivers, and coastal areas in Zhuhai, China. Environ Sci Pollut Res Int 25(26):26209
Li S, Yao Q, Liu J, Wei D, Zhou B, Zhu P, Cui X, Jin J, Liu X, Wang G (2020) Profiles of antibiotic resistome with animal manure application in black soils of northeast China. Journal of Hazardous materials 384:121216
Li, B., Yang, Y., Ma, L., Ju, F., Guo, F., Tiedje, J. and Zhang, T. (2015) Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. Isme Journal.
Lin H, Jiang L, Li B, Dong Y, He Y, Qiu Y (2019) Screening and evaluation of heavy metals facilitating antibiotic resistance gene transfer in a sludge bacterial community. Science of the Total Environment 695:133862
Liu X, Wang H, Zhao H (2020) Propagation of antibiotic resistance genes in an industrial recirculating aquaculture system located at northern China. Environmental Pollution 261:114155
Liu L, Su JQ, Guo Y, Wilkinson DM, Liu Z, Zhu Y-G, Yang J (2018) Large-scale biogeographical patterns of bacterial antibiotic resistome in the waterbodies of China. Environ Int 117(AUG.):292–299
Louca S, Parfrey LW, Doebeli M (2016) Decoupling function and taxonomy in the global ocean microbiome. Science 353(6305):1272–1277
Lu, J., Tian, Z., Yu, J., Yang, M. and Zhang, Y. (2018) Distribution and Abundance of Antibiotic Resistance Genes in Sand Settling Reservoirs and Drinking Water Treatment Plants across the Yellow River, China. Water 10(3), 246-.
Ma L, Li B, Jiang X, Wang Y, Xia Y, Li A, Zhang T (2017) Catalogue of antibiotic resistome and host-tracking in drinking water deciphered by a large scale survey. Microbiome 5(1):154
Marathe NP, Pal C, Gaikwad SS, Jonsson V, Kristiansson E, Larsson D (2017) Untreated urban waste contaminates Indian river sediments with resistance genes to last resort antibiotics. Water Research 124(nov.1):388
Marti E, Variatza E, Balcazar JL (2014) The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol 22(1):36–41
Mazurier S, Merieau A, Bergeau D, Decoin V, Sperandio D, Crépin A, Barbey C, Jeannot K, Vicré-Gibouin M, Plésiat P, Lemanceau P, Latour X (2015) Type III secretion system and virulence markers highlight similarities and differences between human- and plant-associated pseudomonads related to Pseudomonas fluorescens and P. putida. Appl Environ Microbiol 81(7):2579–2590
Miranda CC, de Filippis I, Pinto LH, Coelho-Souza T, Bianco K, Cacci LC, Picão RC, Clementino MM (2015) Genotypic characteristics of multidrug-resistant Pseudomonas aeruginosa from hospital wastewater treatment plant in Rio de Janeiro. Brazil J Appl Microbiol 118(6):1276–1286
Moura A, Soares M, Pereira C, Leitao N, Henriques I, Correia A (2009) INTEGRALL: a database and search engine for integrons integrases and gene cassettes. Bioinformatics 25(8):1096–1098. https://doi.org/10.1093/bioinformatics/btp105
Newman DJ, Cragg GM, Snader KM (2003) Natural Products as Sources of New Drugs over the Period 1981–2002. J Nat Prod 66(7):1022–1037
Peng F, Guo Y, Isabwe A, Chen H, Yang J (2020) Urbanization drives riverine bacterial antibiotic resistome more than taxonomic community at watershed scale. Environment International 137:105524
Qiu Z, Yu Y, Chen Z, Jin M, Yang D, Zhao Z, Wang J, Shen Z, Wang X, Qian D (2012) Nanoalumina promotes the horizontal transfer of multiresistance genes mediated by plasmids across genera. Proc Natl Acad Sci USA 109(13):4944–4949
Servais P, Passerat J (2009) Antimicrobial resistance of fecal bacteria in waters of the Seine river watershed (France). Sci Total Environ 408(2):365–372
Shi W, Zhang H, Li J, Liu Y, Shi R, Du H, Chen J (2019) Occurrence and spatial variation of antibiotic resistance genes (ARGs) in the Hetao Irrigation District, China. Environ Pollut 251:792–801
Siguier P (2006) ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34(90001):D32–D36. https://doi.org/10.1093/nar/gkj014
Wang R, Zhang Y, Cao Z, Wang X, Ma B, Wu W, Hu N, Huo Z, Yuan Q (2019) Occurrence of super antibiotic resistance genes in the downstream of the Yangtze River in China: Prevalence and antibiotic resistance profiles. Sci Total Environ 651(Pt 2):1946–1957
Wen Q, Yang L, Duan R, Chen Z (2016) Monitoring and evaluation of antibiotic resistance genes in four municipal wastewater treatment plants in Harbin. Northeast China Environmental Pollution 212(may):34–40
Xu L, Huang H, Wei W, Zhong Y, Tang B, Yuan H, Zhu L, Huang W, Ge M, Yang S, Zheng H, Jiang W, Chen D, Zhao GP, Zhao W (2014) Complete genome sequence and comparative genomic analyses of the vancomycin-producing Amycolatopsis orientalis. BMC Genomics 15(1):363
Yang Y, Liu W, Xu C, Wei B, Wang J (2017) Antibiotic resistance genes in lakes from middle and lower reaches of the Yangtze River, China: Effect of land use and sediment characteristics. Chemosphere 178:19–25
Yang Y, Li Z, Song W, Du L, Ye C, Zhao B, Liu W, Deng D, Pan Y, Lin H (2019) Metagenomic insights into the abundance and composition of resistance genes in aquatic environments: Influence of stratification and geography. Environ Int 127:371–380
Yang J, Wang H, Roberts DJ, Du HN, Yu XF, Zhu NZ, Meng XZ (2020) Persistence of antibiotic resistance genes from river water to tap water in the Yangtze River Delta. Sci Total Environ 742:140592
Yin X, Jiang XT, Chai B, Li L, Yang Y, Cole JR, Tiedje JM, Zhang T (2018) ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics 34(13):2263–2270
Yuan QB, Zhai YF, Mao BY, Schwarz C, Hu N (2019) Fates of antibiotic resistance genes in a distributed swine wastewater treatment plant. Water Environ Res 91(12):1565–1575
Zgurskaya HI, Rybenkov VV, Krishnamoorthy G, Leus IV (2018) Trans-envelope multidrug efflux pumps of Gram-negative bacteria and their synergism with the outer membrane barrier. Res Microbiol 169(7–8):351–356
Zhang YJ, Hu HW, Yan H, Wang JT, Lam SK, Chen QL, Chen D, He JZ (2019) Salinity as a predominant factor modulating the distribution patterns of antibiotic resistance genes in ocean and river beach soils. Sci Total Environ 668:193–203
Zhang G, Guan Y, Zhao R, Feng J, Huang J, Ma L, Li B (2020) Metagenomic and network analyses decipher profiles and co-occurrence patterns of antibiotic resistome and bacterial taxa in the reclaimed wastewater distribution system. J Hazard Mater 400:123170
Zhang G, Lu S, Wang Y, Liu X, Liu Y, Xu J, Zhang T, Wang Z, Yang Y (2020) Occurrence of antibiotics and antibiotic resistance genes and their correlations in lower Yangtze River. China. Environmental Pollution 257:113365
Zhang G, Lu S, Wang Y, Liu X, Liu Y, Xu J, Zhang T, Wang Z, Yang Y (2020) Occurrence of antibiotics and antibiotic resistance genes and their correlations in lower Yangtze River. China. Environ Pollut 257:113365
Zhao Y, Cocerva T, Cox S, Tardif S, Su JQ, Zhu YG, Brandt KK (2019) Evidence for co-selection of antibiotic resistance genes and mobile genetic elements in metal polluted urban soils. Sci Total Environ 656:512–520
Zheng J, Zhou Z, Wei Y, Chen T, Feng W, Chen H (2018) High-throughput profiling of seasonal variations of antibiotic resistance gene transport in a peri-urban river. Environ Int 114:87–94
Zhou L, Ying G, Zhao J, Yang J, Wang L, Yang B, Liu S (2011) Trends in the occurrence of human and veterinary antibiotics in the sediments of the Yellow River, Hai River and Liao River in northern China. Environ Pollut 159(7):1877–1885
Acknowledgements
We appreciated Xi Zhang and Xiaobo Yang for suggestions for improvement of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (grant no. 51678565), the Special Fund of China (grant no. AWS18J004), andthe Tianjin Natural Science Foundation (grant no. 19JCYBJC23800).
Author information
Authors and Affiliations
Contributions
Chen Zhao and Chenyu Li completed the manuscript. Xiaoming Wang, Zhuosong Cao, Chao Gao, and Sicong Su collected all samples. Bin Xue, Shang Wang, and Zhigang Qiu analyzed the data. Jingfeng Wang and Zhiqiang Shen designed this study.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Robert Duran
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, C., Li, C., Wang, X. et al. Monitoring and evaluation of antibiotic resistance genes in three rivers in northeast China. Environ Sci Pollut Res 29, 44148–44161 (2022). https://doi.org/10.1007/s11356-022-18555-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-022-18555-x