Abstract
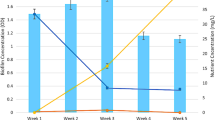
Autotrophic nitrification in biological nitrogen removal systems has been shown to be sensitive to the presence of heavy metals in wastewater treatment plants. Using transcriptase-quantitative polymerase chain reaction (RT-qPCR) data, we examined the effect of copper on the relative expression of functional genes (i.e., amoA, hao, nirK, and norB) involved in redox nitrogen transformation in batch enrichment cultures obtained from a nitrifying bioreactor operated as a continuous reactor (24-h hydraulic retention time). 16S ribosomal RNA (rRNA) gene next-generation sequencing showed that Nitrosomonas-like populations represented 60–70 % of the bacterial community, while other nitrifiers represented <5 %. We observed a strong correspondence between the relative expression of amoA and hao and ammonia removal in the bioreactor. There were no considerable changes in the transcript levels of amoA, hao, nirK, and norB for nitrifying samples exposed to copper dosages ranging from 0.01 to 10 mg/L for a period of 12 h. Similar results were obtained when ammonia oxidation activity was measured via specific oxygen uptake rate (sOUR). The lack of nitrification inhibition by copper at doses lower than 10 mg/L may be attributed to the role of copper as cofactor for ammonia monooxygenase or to the sub-inhibitory concentrations of copper used in this study. Overall, these results demonstrate the use of molecular methods combined with conventional respirometry assays to better understand the response of wastewater nitrifying systems to the presence of copper.




Similar content being viewed by others
References
Ahn JH, Kwan T, Chandran K (2011) Comparison of partial and full nitrification processes applied for treating high-strength nitrogen wastewaters: microbial ecology through nitrous oxide production. Environ Sci Technol 45:2734–2740
Ahn JH, Yu R, Chandran K (2008) Distinctive microbial ecology and biokinetics of autotrophic ammonia and nitrite oxidation in a partial nitrification bioreactor. Biotechnol Bioeng 100:1078–1087
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Barber WP, Stuckey DC (2000) Nitrogen removal in a modified anaerobic baffled reactor (ABR): 2, nitrification. Water Res 34:2423–2432
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522
Çeçen F, Semerci N, Geyik AG (2010) Inhibition of respiration and distribution of Cd, Pb, Hg, Ag and Cr species in a nitrifying sludge. J Hazard Mater 178:619–627
Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, Hauser L, Hooper A, Klotz M, Norton J (2003) Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J Bacteriol 185:2759–2773
Chandran K, Love NG (2008) Physiological state, growth mode, and oxidative stress play a role in Cd (II)-mediated inhibition of Nitrosomonas europaea 19718. Appl Environ Microbiol 74:2447–2453
Chandran K, Smets BF (2000) Applicability of two-step models in estimating nitrification kinetics from batch respirograms under different relative dynamics of ammonia and nitrite oxidation. Biotechnol Bioeng 70:54–64
Chipasa KB (2003) Accumulation and fate of selected heavy metals in a biological wastewater treatment system. Waste Manag 23:135–143
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen A, McGarrell DM, Marsh T, Garrity GM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
Elnabarawy MT, Robideau RR, Beach SA (1988) Comparison of three rapid toxicity test procedures: Microtox,® polytox,® and activated sludge respiration inhibition. Toxicity Assessment 3:361–370
Ensign SA, Hyman MR, Arp DJ (1993) In vitro activation of ammonia monooxygenase from Nitrosomonas europaea by copper. J Bacteriol 175:1971–1980
Francis CA, Beman JM, Kuypers MM (2007) New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J 1:19–27
Gernaey K, Verschuere L, Luyten L, Verstraete W (1997) Fast and sensitive acute toxicity detection with an enrichment nitrifying culture. Water Environ Res 69:1163–1169
Grüttner H, Winther-Nielsen M, Jørgensen L, Bøgebjerg P, Sinkjaer O (1994) Inhibition of the nitrification process in municipal wastewater treatment plants by industrial discharges. Water Sci Technol 29:69–77
Harms G, Layton AC, Dionisi HM, Gregory IR, Garrett VM, Hawkins SA, Robinson KG, Sayler GS (2003) Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ Sci Technol 37:343–351
Hermansson A, Lindgren P-E (2001) Quantification of ammonia-oxidizing bacteria in arable soil by real-time PCR. Appl Environ Microbiol 67:972–976
Hooper AB, Terry KR (1973) Specific inhibitors of ammonia oxidation in Nitrosomonas. J Bacteriol 115:480–485
Hu Z, Chandran K, Grasso D, Smets BF (2002) Effect of nickel and cadmium speciation on nitrification inhibition. Environ Sci Technol 36:3074–3078
Hu Z, Chandran K, Grasso D, Smets BF (2003) Impact of metal sorption and internalization on nitrification inhibition. Environ Sci Technol 37:728–734
Hu Z, Chandran K, Grasso D, Smets BF (2004) Comparison of nitrification inhibition by metals in batch and continuous flow reactors. Water Res 38:3949–3959
Jönsson K, Grunditz C, Dalhammar G, La Cour JJ (2000) Occurrence of nitrification inhibition in Swedish municipal wastewaters. Water Res 34:2455–2462
Juliastuti S, Baeyens J, Creemers C, Bixio D, Lodewyckx E (2003) The inhibitory effects of heavy metals and organic compounds on the net maximum specific growth rate of the autotrophic biomass in activated sludge. J Hazard Mater 100:271–283
Kapoor V, Pitkänen T, Ryu H, Elk M, Wendell D, Santo Domingo JW (2015) Distribution of human-specific bacteroidales and fecal indicator bacteria in an urban watershed impacted by sewage pollution, determined using RNA-and DNA-based quantitative PCR assays. Appl Environ Microbiol 81:91–99
Karvelas M, Katsoyiannis A, Samara C (2003) Occurrence and fate of heavy metals in the wastewater treatment process. Chemosphere 53:1201–1210
Kim YM, Lee DS, Park C, Park D, Park JM (2011) Effects of free cyanide on microbial communities and biological carbon and nitrogen removal performance in the industrial activated sludge process. Water Res 45:1267–1279
Kim YM, Park D, Lee DS, Park JM (2008) Inhibitory effects of toxic compounds on nitrification process for cokes wastewater treatment. J Hazard Mater 152:915–921
Kuo DH-W, Robinson KG, Layton AC, Meyers AJ, Sayler GS (2010) Transcription levels (amoA mRNA-based) and population dominance (amoA gene-based) of ammonia-oxidizing bacteria. J Ind Microbiol Biotechnol 37:751–757
Madoni P, Davoli D, Guglielmi L (1999) Response of SOUR and AUR to heavy metal contamination in activated sludge. Water Res 33:2459–2464
Miller DJ, Nicholas D (1985) Characterization of a soluble cytochrome oxidase/nitrite reductase from Nitrosomonas europaea. J Gen Microbiol 131:2851–2854
Milner M, Curtis T, Davenport R (2008) Presence and activity of ammonia-oxidising bacteria detected amongst the overall bacterial diversity along a physico-chemical gradient of a nitrifying wastewater treatment plant. Water Res 42:2863–2872
Ochoa-Herrera V, León G, Banihani Q, Field JA, Sierra-Alvarez R (2011) Toxicity of copper (II) ions to microorganisms in biological wastewater treatment systems. Sci Total Environ 412:380–385
Painter HA (1970) A review of literature on inorganic nitrogen metabolism in microorganisms. Water Res 4:393–450
Park S, Ely RL (2008) Candidate stress genes of Nitrosomonas europaea for monitoring inhibition of nitrification by heavy metals. Appl Environ Microbiol 74:5475–5482
Radniecki T, Ely R (2008) Zinc chloride inhibition of Nitrosococcus mobilis. Biotechnol Bioeng 99:1085–1095
Radniecki TS, Semprini L, Dolan ME (2009) Expression of merA, amoA and hao in continuously cultured Nitrosomonas europaea cells exposed to zinc chloride additions. Biotechnol Bioeng 102:546–553
Rowan AK, Snape JR, Fearnside D, Barer MR, Curtis TP, Head IM (2003) Composition and diversity of ammonia‐oxidising bacterial communities in wastewater treatment reactors of different design treating identical wastewater. FEMS Microbiol Ecol 43:195–206
Sakano Y, Pickering KD, Strom PF, Kerkhof LJ (2002) Spatial distribution of total, ammonia-oxidizing, and denitrifying bacteria in biological wastewater treatment reactors for bioregenerative life support. Appl Environ Microbiol 68:2285–2293
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Siripong S, Rittmann BE (2007) Diversity study of nitrifying bacteria in full-scale municipal wastewater treatment plants. Water Res 41:1110–1120
Walker C, De La Torre J, Klotz M, Urakawa H, Pinel N, Arp D, Brochier-Armanet C, Chain P, Chan P, Gollabgir A (2010) Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci U S A 107:8818–8823
You S, Hsu C, Ouyang C (2002) Identification of the microbial diversity of wastewater nutrient removal processes using molecular biotechnology. Biotechnol Lett 24:1361–1366
You S-J, Tsai Y-P, Huang R-Y (2009) Effect of heavy metals on nitrification performance in different activated sludge processes. J Hazard Mater 165:987–994
Yu R, Chandran K (2010) Strategies of Nitrosomonas europaea 19718 to counter low dissolved oxygen and high nitrite concentrations. BMC Microbiol 10:70
Yu R, Lai B, Vogt S, Chandran K (2011) Elemental profiling of single bacterial cells as a function of copper exposure and growth phase. PLoS One 6:e21255
Acknowledgments
We thank Kit Daniels for building the nitrifying bioreactor and for technical assistance. VK and XL were supported by ORISE-EPA Research Fellowship. KC was supported by the Water Environment Research Foundation. The US Environmental Protection Agency, through its Office of Research and Development, funded and managed, or partially funded and collaborated in, the research described herein. This work has been subjected to the agency’s administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Gerald Thouand
Rights and permissions
About this article
Cite this article
Kapoor, V., Li, X., Chandran, K. et al. Use of functional gene expression and respirometry to study wastewater nitrification activity after exposure to low doses of copper. Environ Sci Pollut Res 23, 6443–6450 (2016). https://doi.org/10.1007/s11356-015-5843-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-015-5843-2