Abstract
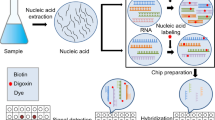
Warning and monitoring projects of harmful algal blooms require simple and rapid methods for simultaneous and accurate detection and identification of causative algae present in the environmental samples. Here, reverse dot blot hybridization (RDBH) was employed to simultaneously detect several harmful algae by using five representative bloom-forming microalgae along the Chinese coast. A set of specific probes for RDBH were developed by PCR, cloning, and sequencing of the internal transcribed spacer (ITS), alignment analysis, and probe design. Each probe was oligo (dT)-tailed and spotted onto positively charged nylon membrane to make up a low-density oligonucleotide array. Universal primers designed within the conserved regions were used to amplify the ITS sequences by using genomic DNA of target as templates. The digoxigenin (Dig)-labeled PCR products were denatured and then hybridized to the oligonucleotide array. The array produced a unique hybridization pattern for each target species differentiating them from each other. The preparations of oligonucleotide array and hybridization conditions were optimized. The developed RDBH demonstrated a detection limit up to 10 cells. The detection performance of RDBH was relatively stable and not affected by non-target species and the fixation time of target species over at least 30 days. The RDBH could recover all the target species from the simulated field samples and target species confirmed by the subsequent microscopy examination in the environmental samples. These results indicate that RDBH can be a new technical platform for parallel discrimination of harmful algae and is promising for environmental monitoring of these microorganisms.







Similar content being viewed by others
References
Ahn S, Kulis DM, Erdner DL, Anderson DM, Walt DR (2006) Fiber-optic microarray for simultaneous detection of multiple harmful algal bloom species. Appl Environ Microbiol 72:5742–5749
Chen GF, Liu Y, Zhang CY, Ma CS, Zhang BY, Wang GC (2013) Development of rRNA-targeted probes for detection of Prorocentrum micans (Dinophyceae) using whole cell in situ hybridization. J Appl Phycol 25:1077–1089
Connell LB (2001) Nuclear ITS region of the alga Heterosigma akashiwo (Chromophyta: Raphidophyceae) is identical in isolates from Atlantic and Pacific basins. Mar Biol 160:953–960
Diercks S, Metfies K, Medlin LK (2008) Development and adaptation of a multiprobe biosensor for the use in a semi-automated device for the detection of toxic algae. Biosens Bioelectron 23:1527–1533
Galluzzi L, Cegna A, Casabianca S, Penna A, Saunders N, Magnani M (2011) Development of an oligonucleotide microarray for the detection and monitoring of marine dinoflagellates. J Microbiol Methods 84:234–242
Guillard RRL (1975) Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH (eds) Culture of marine invertebrate animals. Plenum, New York, pp 29–60
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Kegel JU, Amo YD, Medlin LK (2013) Introduction to project MIDTAL: its methods and samples from Arcachon Bay, France. Environ Sci Pollut Res 20:6690–6704
Ki JS, Han MS (2006) A low-density oligonucleotide array study for parallel detection of harmful algal species using hybridisation of consensus PCR products of LSU rDNA D2 domain. Biosens Bioelectron 21:1812–1821
Kodani M, Mixson-Hayden T, Drobeniuc J, Kamili S (2014) Rapid and sensitive approach to simultaneous detection of genomes of hepatitis A, B, C, D and E viruses. J Clin Virol 61:260–264
Medlin LK (2013) Note: steps taken to optimise probe specificity and signal intensity prior to field validation of the MIDTAL (Microarray for the Detection of Toxic Algae). Environ Sci Pollut Res 20:6686–6689
Mikulski CM, Park YT, Jones KL, Lee CK, Lim WA, Lee Y, Scholin CA, Doucette GJ (2008) Development and field application of rRNA-targeted probes for the detection of Cochlodinium polykrikoides Margalef in Korean coastal waters using whole cell and sandwich hybridization formats. Harmful Algae 7:347–359
Penna A, Galluzzi L (2013) The quantitative real-time PCR applications in the monitoring of marine harmful algal bloom (HAB) species. Environ Sci Pollut Res 20:6851–6862
Peplies J, Glockner O, Amann R (2003) Optimization strategies for DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl Environ Microbiol 69:1397–1407
Schlötterer C, Hauser MY, von Haeseler A, Tautz D (1994) Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Mol Biol Evol 11:513–522
Scholin CA, Buck KR, Britschgi T, Cangelosi G, Chavez FP (1996) Identification of Pseudo-nitzschia australis (Bacillariophyceae) using rRNA-targeted probes in whole cell and sandwich hybridization formats. Phycologia 35:190–197
Scorzetti G, Brand LE, Hitchcock GL, Rein KS, Sinigalliano CD, Fell JW (2009) Multiple simultaneous detection of harmful algal blooms (HABs) through a high throughput bead array technology, with potential use in phytoplankton community analysis. Harmful Algae 8:196–211
Sherlock G (2000) Analysis of large-scale gene expression data. Curr Opin Immunol 12:201–205
Sieburth JM, Johnson PW, Hargraves PE (1988) Ultrastructure and ecology of Aureococcus anophagefferens gen-et-sp-nov (Chrysophyceae)—the dominant picoplankter during a bloom in Narragansett Bay, Rhode-Island, summer 1985. J Phycol 24:416–425
Steane DA, Mcclure BA, Clarke AE, Kraft GT (1991) Amplification of the polymorphic 5.8S rRNA gene from selected Australian gigartinalean species (Rhodophyta) by polymerase chain reaction. J Phycol 27:758–762
Sun ZZ, Peng Y, Zhang MC, Wang K, Bai JL, Li XL, Ning BA, Gao ZX (2014) Simultaneous and highly sensitive detection of six different foodborne pathogens by high-throughput suspension array technology. Food Control 40:300–309
Taylor JD, Berzano M, Percy L, Lewis J (2013) Evaluation of the MIDTAL microarray chip for monitoring toxic microalgae in the Orkney Islands, U.K. Environ Sci Pollut Res 20:6765–6777
Wang HX, Lu DD, Huang HY, Dai XF, Xia P (2012) Morphological and phylogenetic analysis of Karlodinium veneficum isolated from the East China Sea in China. Chin Bull Bot 461:179–188
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand J, Sainsky J, White T (eds) PCR protocols: a guide to methods and applications. Academic, Orlando, Florida, pp 315–322
Ye RW, Wang T, Bedzyk L, Croker KM (2001) Applications of DNA microarrays in microbial systems. J Microbiol Methods 47:257–272
Zhang QC, Qiu LM, Yu RC, Kong FZ, Wang YF, Yan T, Gobler CJ, Zhou MJ (2012) Emergence of brown tides caused by Aureococcus anophagefferens Hargraves et Sieburth in China. Harmful algae 19:117–124
Zhen Y, Mi TZ, Yu ZG (2009) Detection of several harmful algal species by sandwich hybridization integrated with a nuclease protection assay. Harmful Algae 8:651–657
Zhou MJ, Zhu MY, Zhang J (2001) Status of harmful algal blooms and related research activities in China. Chin Bull Life Sci 13:54–59
Zingone A, Percopo I, Sims PA, Sarno D (2005) Diversity in the genus Skeletonema (Bacillariophyceae). I. A reexamination of the type material of S. costatum with the description of S. grevillei sp. nov. J Phycol 41:140–150
Acknowledgments
This work was supported by the National Scientific Foundation of China (41476086, 41106082, 41176141); the Open Fund of Key Laboratory of Marine Ecology and Environmental Science, Institute of Oceanology, Chinese Academy of Sciences (KLMEES201303); and Basic Research of Harbin Institute of Technology Outstanding Talents Cultivation Plan of Class III.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 179 kb)
Rights and permissions
About this article
Cite this article
Chen, G.F., Zhang, C.Y., Wang, Y.Y. et al. Application of reverse dot blot hybridization to simultaneous detection and identification of harmful algae. Environ Sci Pollut Res 22, 10516–10528 (2015). https://doi.org/10.1007/s11356-015-4141-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-015-4141-3