Abstract
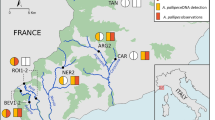
Rhodeus atremius suigensis is an extremely endangered bitterling fish, designated as one of the Nationally Endangered Species of Wild Fauna and Flora by the Ministry of Environment of Japan. To prevent its extinction, effective ecological survey methods are necessary in order to guide habitat management. Recently, environmental DNA (eDNA) analysis has proven useful for detection and censusing of endangered aquatic species that are difficult to locate and capture. In this study, we developed and evaluated a semi-quantitative system to track R. a. suigensis using eDNA. We designed primers and a fluorescent probe specific to the mitochondrial gene for NADH dehydrogenase subunit 1 (ND-1) of R. a. suigensis for quantitative real-time PCR. We first confirmed the utility of this analysis in aquarium experiments, before performing field surveys. We sampled water for eDNA analysis and set fish traps at 48 points in an agricultural channel in the Ashida River basin, inhabited by R. a. suigensis, and examined the relationship between fish occurrence and eDNA concentration. The concentration was correlated with the distance downstream from the point where specimens of R. a. suigensis were captured, indicating that the eDNA concentration reflects distribution and abundance of R. a. suigensis. These results not only demonstrate the reliability of this method, but also suggest its feasibility for conservation measures.






Similar content being viewed by others
Data availability
All the qPCR results used in this study are shown in Supplementary Table 1.
References
Abe T, Onikura T, Nakajima A (2015) Current status and conservation problems of two Rhodeus fishes in Japan. Jpn J Ichthyol 62(1):65–69 (in Japanese)
Akamatsu Y, Inui R, Hitotsumatsu A, Kono T, Doi H (2017) Fundamental study on estimating abundance of stream fish using environmental DNA analysis. J Jpn Soc Civil Eng Ser B1 73(4):I_1111-I_1116 (in Japanese with English abstract)
Bohmann K, Evans A, Gilbert MTP, Carvalho GR, Creer S, Knapp M, Yu DW, de Bruyn M (2014) Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol Evol 29(6):358–367
Doi H, Inui R, Akamatsu Y, Kanno K, Yamanaka H, Takahara T, Minamoto T (2017) Environmental DNA analysis for estimating the abundance and biomass of stream fish. Freshw Biol 62:30–39
Ficetola GF, Miaud C, Pompanon F, Taberlet P (2008) Species detection using environmental DNA from water samples. Biol Lett 4(4):423–425
Fukumoto S, Ushimaru A, Minamoto T (2015) A basin-scale application of environmental DNA assessment for rare endemic species and closely related exotic species in rivers: a case study of giant salamanders in Japan. J Appl Ecol 52(2):358–365
Jo T, Yamanaka H (2022) Meta-analyses of environmental DNA downstream transport and deposition in relation to hydrogeography in riverine environments. Freshw Biol 67:1333–1334
Kawamura K (2014) Rhodeus atremius suigensis. In: Ministry of the Environment Government of Japan (ed) Red data book 2014, brackish and fresh water fishes. Gyosei, Tokyo, pp 30–31 (in Japanese)
Kitamura J, Uchiyama R (2020) Bitterling fishes of Japan: natural history and culture. Yama-Kei, Tokyo (in Japanese)
Klymus KE, Merkes CM, Allison MJ, Goldberg CS, Helbing CC, Hunter ME, Jackson CA, Lance RF, Mangan AM, Monroe EM, Piaggio AJ, Stokdyk JP, Wilson CC, Richter CA (2020) Reporting the limits of detection and quantification for environmental DNA assays. Environ DNA 2:271–282
Koizumi N, Minamoto T, Shirako T, Nakamura M (2022) Applicability of environmental DNA research and diffusion distance of environmental DNA in agricultural canals. Water, Land Environ Eng 90(8):571–574 (in Japanese)
Laporte M, Bougas B, Côté G, Champoux O, Paradis Y, Morin J, Bernatchez L (2020) Caged fish experiment and hydrodynamic bidimensional modeling highlight the importance to consider 2D dispersion in fluvial environmental DNA studies. Environ DNA 2:362–372
Minamoto T, Miya M, Sado T, Seino S, Doi H, Kondoh M, Nakamura K, Takahara T, Yamamoto S, Yamanaka H, Araki H, Iwasaki W, Kasai A, Masuda R, Uchii K (2021) An illustrated manual for environmental DNA research: water sampling guidelines and experimental protocols. Environ DNA 3:8–13
Mizumoto H, Misuzuka T, Araki H (2020) An environmental DNA survey on distribution of an endangered salmonid species, Parahucho perryi, in Hokkaido. Japan Front Ecol Evol 8:569425
Nakata K, Kobayashi S, Kawamoto I, Miyatake Y, Aoe H (2017a) Utilization of artificial spawning beds by endangered bittering fish in an agricultural channel in southern Okayama, western Japan. Ecol Civil Eng 20(1):33–41 (in Japanese with English abstract)
Nakata K, Miyatake Y, Kawai K, Kobayashi S, Ham SN, Saito M, Aoe H (2017b) Habitat preference by the endangered bitterling fish Rhodeus atremius suigensis in an agricultural channel in southern Okayama, western Japan. Ecol Civil Eng 19(2):117–130 (in Japanese with English abstract)
Nakata K, Miyoshi Y, Takeuchi K, Koizumi N, Nakamura M (2019) Distribution of freshwater mussel species in a habitat of an endangered bittering species in the Ashida River system, Hiroshima Prefecture, western Japan. Advance Abstracts for the 52nd Annual Meeting, 2019: 131 (in Japanese)
Nukazawa K, Hamasuna Y, Suzuki Y (2018) Simulating the advection and degradation of the environmental DNA of common carp along a river. Environ Sci Technol 52:10562–10570
Saitoh K (2021) Conservation issues partly solved on “Suigenzenitanago” (Rhodeus smithii complex) impaired by the taxonomic confusion. Ichthyol Res 68:322–324
Sakata MK, Miki N, Sugiyama H, Minamoto T (2017) Identifying a breeding habitat of a critically endangered fish, Acheilognathus typus, in a natural river in Japan. Sci Nat 104:100
Thomsen FP, Willerslev E (2015) Environmental DNA – An emerging tool in conservation for monitoring past and present biodiversity. Biol Conserv 183:4–18
Yamanaka H, Minamoto T, Matsuura J, Sakurai S, Tsuji S, Motozawa H, Hongo M, Sogo Y, Kakimi N, Teramura I, Sugita M, Baba M, Kondo A (2017) A simple method for preserving environmental DNA in water samples at ambient temperature by addition of cationic surfactant. Limnology 18:233–241
Acknowledgements
We thank Mr. Satoshi Furumoto, Mr. Hidenori Yoshigou and Dr. Shigefumi Kanao for providing information on the previous distribution of Rhodeus atremius suigensis in the Ashida River basin. We also thank Mr. Kazuya Takeuchi (Okayama University) and the staff of the Environment Conservation Division, City of Fukuyama for their help in our field investigations. We are grateful to Dr. Risa Monna (Rizo Inc.) for her advice on this research. We thank Dr. Steven D. Aird for editing the manuscript. This study was financially supported by the Chugoku Regional Development Bureau, Ministry of Land, Infrastructure, Transport and Tourism of Japan and by the Suma Aqualife Park, Kobe City. This study complies with current Japanese law.
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
11355_2022_531_MOESM2_ESM.pptx
Supplementary Fig. 1. Rhodeus atremius suigensis eDNA concentration is not correlated with water depth or velocity. Water depth (A)/water velocity (B) (horizontal axis) and eDNA concentration (vertical axis) at each sampling site are plotted. There was no significant rank correlation between water depth or water velocity and eDNA concentration (Kendall rank correlation coefficient, water depth: τ = 0.05, P > 0.05, flow velocity: τ = 0.03, P > 0.05) Supplementary file2 (PPTX 517 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Otsuki, K., Hamada, M., Koizumi, N. et al. Quantitative PCR method to detect an extremely endangered bitterling fish (Rhodeus atremius suigensis) using environmental DNA. Landscape Ecol Eng 19, 79–86 (2023). https://doi.org/10.1007/s11355-022-00531-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11355-022-00531-9