Abstract
The Amazonian cacao tree or the chocolate tree (Theobroma cacao L.) is cultivated to produce seeds, the valuable raw material for the chocolate industry. However, cacao yield is hampered by diseases and low productivity. Cacao single nucleotide polymorphism (SNP) markers associated with breeding and adaptive traits provide a genetic tool for improving selection and reducing breeding cycles. Here, we genotyped 40 published SNPs associated with disease resistance and 11 SNPs with yield traits in 346 accessions using the MassARRAY® system. These 51 SNPs were identified in different breeding experiments in Costa Rica, Brazil, and Trinidad and Tobago. Four genetic clusters were identified, and two of them were observed in high proportion in managed cacao, characterized by a high proportion of admixed individuals reflecting the man-made hybrids. One cluster overrepresented in managed cacao is associated with the preference to keep superior flavor in cultivated cacao from Criollo ancestry. Introgression of wild material collected in Peru associated with disease resistance to witches’ broom disease (WBD) was also observed. Underutilized genetic resources were observed in managed cacao such as the Guiana cacao, while previous agronomic evaluation has demonstrated its good yield potential. Yield and disease resistance traits (mainly resistance to WBD) show divergence between wild and managed cacao (FST > 0.05) probably reflecting selection during domestication, cultivation, and breeding efforts. Eleven SNPs showed a divergence between wild and managed cacao and can be used to build breeding populations. New collections of wild cacao followed by agronomic evaluations can broaden the genetic base of cultivated cacao, especially for such traits as disease resistance to frosty pod rot (FPR).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cacao tree (Theobroma cacao L.) is a valuable crop tree commonly cultivated in the tropics to produce seeds, the raw material that sustains the global chocolate industry (Coe and Coe 2013; Dand 2011). The putative origin of cacao is western Amazonia (Upper Amazon basin), and its natural range includes the wide Amazon basin where the primary gene pool of cacao is structured in twelve genetic clusters (Gutiérrez et al. 2021a; Motamayor et al. 2008; Nieves-Orduña et al. 2021, 2023; Sereno et al. 2006; Zhang et al. 2011, 2012). Seven of these genetic clusters were identified in natural populations in western Amazonia, at the Ecuadorian-Peruvian Amazon, whereas one cluster is dominant in eastern Amazonia and French Guiana (Cornejo et al. 2018; Motamayor et al. 2008). Further population genetic analysis on areas not yet studied can identify new clusters and potential genetic resources for breeding (Nieves-Orduña et al. 2023). Cacao populations from western Amazonia were likely spread by humans to Eastern Amazonia (Levis et al. 2017). This dispersal generated a gradient of cacao genetic diversity declining from west to east Amazonia (Cornejo et al. 2018; Motamayor et al. 2008; Nieves-Orduña et al. 2021). New evidence based on ancient DNA suggests that admixed cacao genotypes experienced human dispersal from the Peruvian Amazon and adaptation to new environments in Coastal Ecuador and North of Colombia (Lanaud et al. 2024). In addition, the influences of paleoclimates and forest refugial areas also shaped the distribution of cacao in Amazonia (Lachenaud 1997; Motamayor et al. 2008; Thomas et al. 2012).
The cacao genetic resources are conserved mainly in two international and several national collections (Bekele and Phillips-Mora 2019). Continued deforestation in wild habitats threatens wild cacao diversity if proper in situ conservation and new collections to enrich current ex situ collections are not implemented (Nieves-Orduña et al. 2023). The International Cocoa Quarantine Centre at the University of Reading (ICQC, R) in the UK facilitates the global distribution of pathogen-free plant material (Daymond 2018), but cultivated cacao represents only a fraction of the species’ genetic diversity (Bennett 2003; Boza et al. 2014; Zhang et al. 2011; Zhang and Motilal 2016). The global demand for chocolate has increased cacao cultivation, but it is hampered by diseases and low yield (Gutiérrez et al. 2016; Ploetz 2016). In addition, there is evidence of deforestation linked to cacao expansion mainly in west Africa (Hoang and Kanemoto 2021; Kalischek et al. 2023) but also increased forest cover through agroforestry (Orozco-Aguilar et al. 2021). Compared to advances in the productivity of tropical crops, such as oil palm, average global cacao production has remained low since 1961 (Morrissey et al. 2019).
Advances in cacao molecular marker-assisted breeding and genomic selection can improve breeding populations and help to select high-yield and disease-resistant cacao genotypes (Schnell et al. 2007). Still, cacao breeding depends on the extensive genetic variation existing in wild populations present in Amazonia (Motamayor et al. 2008; Nieves-Orduña et al. 2023). Genome-wide association studies (GWAS) and quantitative trait locus (QTL) mapping in cacao have identified single nucleotide polymorphism (SNP) markers linked to resistance to the black pod rot (BPR) disease caused by Phytophthora spp. (Gutiérrez et al. 2021b), witches’ broom disease (WBD) caused by Moniliophthora perniciosa (Motilal et al. 2016; Royaert et al. 2016), Ceratocystis wilt (CW) caused by Ceratocystis cacaofunesta (Fernandes et al. 2018), and moniliasis or frosty pod rot (FPR) caused by Moniliophthora roreri (Gutiérrez et al. 2021b). In addition, SNPs linked to high-yield traits and sexual compatibility were identified (da Silva et al. 2016; Fernandes et al. 2020). Thus, favorable identified genetic variation can be exploited for improving the selection efficiency and reducing breeding cycles.
We present here our research aimed to characterize a diverse set of 346 reference cacao accessions representing both wild and managed cacao genotyped at 51 published SNPs associated with important cacao agronomic traits, such as disease resistance, yield, and sexual compatibility, using the MassARRAY® system (Agena Bioscience, Hamburg, Germany). Specifically, our aims were to (1) describe the population structure and analyze genetic clusters in wild and managed cacao, (2) identify new genetic resources for cacao breeding based on SNP profiles, and (3) identify signatures of selection for agronomic traits that differentiate wild and managed cacao to assist breeding of superior cacao genotypes. Using published phenotypic data, we aimed to also validate disease resistance and yield associated SNP alleles, and uncover valuable cacao accessions and new genetic resources to be used in breeding efforts. The obtained SNP profiles will be reported to the International Cocoa Germplasm Database (ICGD). The accessions analyzed here are in the public domain and can be accessed by any cacao-producing country, facilitating the validation of results through breeding programs and farm conditions.
Materials and methods
Germplasm and DNA isolation
The germplasm analyzed consisted of 346 cacao accessions collected in wild habitats and from managed cacao. The managed cacao included 168 clones representing different cultivars, selections, and breeding populations from several groups such as Refractario, Estación Experimental Tropical (EET), United Fruit (UF), Imperial College Selection (ICS), Trinidad Select Hybrid (TSH), SIAL (Selecao Instituto Agronomico do Leste), Selecao Instituto do Cacau (SIC), and Tropical Agriculture Research Service (TARS-Series of cacao) (Turnbull and Hadley 2023). The wild germplasm samples included 178 accessions, representing mainly northwestern Amazonia (77% of the samples), which is considered the hot spot of cacao genetic diversity (Clement et al. 2010; Cornejo et al. 2018), and French Guiana (23%). The wild cacao germplasm represented a wide geographic distribution within the cacao primary gene pool, including samples from five countries, mostly from Peru (57%). They included accessions known as the Pound Collection composed of groups coded as IMC, MO, NA, PA, POUND, and SCA. These accessions were collected in the 1930s by Frederick J. Pound in the Peruvian Amazon while searching for genotypes resistant to WBD that are now the basis for breeding WBD-resistant cacao (Bartley 2005; Zhang et al. 2011). More recent collections included cacao accessions coded as LCT EEN collected in the Ecuadorian Amazon (Allen 1988) and GU accessions from French Guiana (Lachenaud 2015). Although collected in the wild as seeds or budwood, these groups of accessions may have experienced human intervention such as cultivation or translocation, considering the persistent effects of pre-Columbian societies in plant domestication in Amazonia (Barlow et al. 2012; Clement et al. 2015; Levis et al. 2017). Among the accessions analyzed, 137 clones represented the ten genetic groups identified by Motamayor et al. (2008): Criollo (3), Amelonado (22), Nacional, which represent, traditional cultivars (2), Curaray (10), Contamana (5), Guiana (15), Iquitos (30), Marañon (32), Nanay (10), and Purus (8). The complete list of samples studied is presented in Supplementary Table S1. In addition, the ICGD (http://www.icgd.rdg.ac.uk) provides updated agronomic details, geographic origin, and passport data of the germplasm analyzed (Turnbull and Hadley 2023).
The 346 cacao accessions analyzed are subject to international distribution under the International Treaty on Plant Genetic Resources for Food and Agriculture (CacaoNet 2012) and were obtained thanks to the ICQC at the University of Reading, UK, and the germplasm cacao collection held in the International Center for Tropical Agriculture (CATIE) in Costa Rica. We used 1 cm2 of fresh leaf tissue per sample and the DNeasy 96 Plant Kit (Qiagen, Hilden, Germany) for DNA extraction.
Selection of the SNP markers
We selected 51 cacao SNPs from the published data that are supposedly associated with important agronomic traits such as disease resistance (40 SNPs) and yield (11 SNPs), and genotyped them in wild and managed cacao accessions in our study (Table 1). This set of SNPs was identified by GWAS and QTL analysis in cacao research centers at Trinidad (Motilal et al. 2016), Costa Rica (Gutiérrez et al. 2021b), and Brazil (da Silva et al. 2016; Fernandes et al. 2018, 2020; Royaert et al. 2016). The SNP panel included eight SNPs associated with disease resistance to BPR (Gutiérrez et al. 2021b), 11 SNPs linked to WBD resistance (Motilal et al. 2016; Royaert et al. 2016), 16 SNPs related to FPR resistance (Gutiérrez et al. 2021b), and five SNPs linked to CWC resistance (Fernandes et al. 2018). In addition, the SNP set included three SNPs associated with a number of seeds (Motilal et al. 2016), four SNPs related to yield components such as dry seed weight, number of pods harvested, average yield, and high pod index, respectively (Fernandes et al. 2020), and four SNPs associated with flower retention (as a measure of self-compatibility) (da Silva et al. 2016).
Genotyping with MassARRAY system
The SNPs’ flanking sequences were obtained from the published data (see Table 1) and were used to design two SNP assays for the MassARRAY® system using the Assay Design Suite V2.0 (Agena Bioscience 2015). Primer adjustment, PCR amplification, SAP treatment, and iPLEX reaction were done following the instructions of the manufacturer (Agena Bioscience 2019a). Allele calling was conducted using Typer Analyzer v.5.0.2137 (Agena Bioscience 2019b). SNPs with at least 75% genotyping success call rate across all samples were retained for further analysis, and the same genotyping success call rate threshold was used for all DNA samples. Thus, the final data set for further analysis included 318 DNA samples and 42 SNPs (Supplementary Tables S1, S2, and S3).
Data analysis
Analysis of molecular variance (AMOVA) and FST between wild and managed cacao, FST per locus, and principal coordinate analysis (PCoA) were performed in GenAlEx 6.5 using 999 permutations (Peakall and Smouse 2012). Analysis of population structure and admixture was done using STRUC-TURE 2.3.4 (Pritchard et al. 2000) with the admixture model and correlated allele frequencies of 42 SNPs by testing from K = 1 to K = 10 subpopulations with 10 repetitions for each K. The numbers of burn-ins and iterations were 10,000 and 100,000, respectively. Structure Harvester 0.6.94 (Earl and vonHoldt 2012) was used to determine the most likely number of clusters (K) using the delta K Evanno method, and obtained results were visualized using CLUMPAK (Kopelman et al. 2015).
An UPGMA dendrogram based on the pairwise Nei’s standard genetic distance matrix of individuals genotyped for 42 SNPs was generated using Populations 1.2.32 (Langella 2001) with 999 bootstraps on loci and visualized using Interactive Tree of Life (iTOL) 6.7.4 (Letunic and Bork 2021).
Phenotypic trait data to validate favorable SNPs associated with agronomic traits were collected from published data and the International Cocoa Germplasm Data Base (Turnbull and Hadley 2023). Based on these published data, accessions were described as either susceptible or tolerant/resistant, while no clear distinction was made between the terms tolerant and resistant. Fisher’s exact test was calculated in Statistica (StatSoft Europe GmbH, Hamburg, Germany) to compare genotype distributions of wild and managed cacao, and of resistant-tolerant and susceptible germplasm to WBD and CW. Relative genotype frequencies per SNP were calculated in Excel (Microsoft Corporation, Redmond, Washington, USA). The map showing the geographic distribution of identified cacao clusters was developed using ArcGIS software (www.esri.com).
Results
Genetic structure of accessions observed in wild and managed cacao
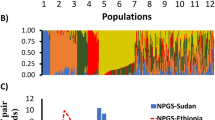
Although the structure analysis did not resolve the exact ten genetic clusters identified earlier in cacao populations based on microsatellite markers (Motamayor et al. 2008), it suggests four most likely clusters (K = 4) for the studied accessions based on 42 SNPs (Fig. 1 and Supplementary Figs. S1 and S2).
Genetic admixture of four clusters identified in wild and managed cacao accessions based on 42 SNPs. a geographic distribution of clusters 1–4 in wild cacao. b Q-values of individual wild cacao accessions arranged along their geographical location from west to east Amazonia. c Q-values of individual managed cacao accessions arranged from high to low admixture of the second cluster. MO (Morona), IMC (Iquitos Mixed Calabacillo), NA (Nanay), POUND, PA (Parinari), and SCA (Scavina) are the names of groups of accessions from the Pound Collection
We observed a clear pattern of differentiation of the wild germplasm when it was arranged from west to east Amazonia. Accessions collected in the Ecuadorian Amazon showed a high admixture proportion of cluster two (Fig. 1A). Cluster one was observed in high proportion in the Peruvian Amazon. The IMC, NA, and PA germplasm series collected around Iquitos, Peru, have similar structure and are dominated by cluster one, but the PA series also admixed much with the fourth cluster (Fig. 1B). The MO series collected in Peru has a high admixture from the third cluster, which has a high representation in managed cacao. The SCA series shows a high proportion of the second cluster. The accessions collected in French Guiana can be distinguished by a high admixture from the fourth cluster (Fig. 1). No private alleles were observed in the data set, likely due to the limited number of samples and SNPs studied here.
We observed that managed cacao accessions have mainly admixture from the second and third clusters, with a little admixture from the first cluster (Fig. 1). A high proportion of the second cluster was observed in the Criollo accessions (Criollo 12, 13, 65), which represent the first domesticated cacao (Cornejo et al. 2018). A higher proportion of the third cluster was observed in clones developed in Brazil, such as SIAL and SIC. In addition, we identified underrepresentation of the fourth cluster associated with Guiana accessions. Overall, managed cacao includes largely admixed accessions, reflecting hybrids in breeding populations and cultivars such as UF, ICS, TARS, CCN 51, and CATIE R6 (Fig. 1 and Supplementary Fig. 2B).
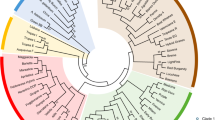
Although there is an overlap between wild and managed cacao in the PCoA, we observed some clustering trends in wild cacao and managed cacao (Fig. 2). Geographically, accessions collected in eastern Amazonia, GU-Guiana, are clustered in the lower left of the PCoA. While wild accessions collected in western Amazonia tend to be clustered on the right side of the PCoA, accessions such as SCA collected in central Peru are located in the lower right of the PCoA (Fig. 2). The dendrogram shows a similar pattern as the PCoA, the Guiana accessions are separated as a distinct group, sister to another group represented by a subset of accessions of managed cacao. The rest of the accessions of managed cacao are not well-resolved in the dendrogram (Fig. 3).
Principal coordinates analysis (PCoA) of 318 cacao accessions based on genetic distance matrix estimated with 42 SNP markers linked to agronomic traits. The first component of the PCoA explains 13.09% of the total variation, and the second component explains 8.31%. Different dot colors represent different wild accessions; open green circles depict managed cacao accessions
Managed cacao accessions tend to be clustered in the upper left of the PCoA, specially selected cacao such as SIC and SIAL developed in Bahia, Brazil, but Criollo accessions (Criollo 12, 13, 65) are clustered on the right side. Within the managed cacao, cultivars and selections of high use in breeding programs such as CCN 51 and UF 273 type 1 are located in the center of the PCoA. CCN 51 is plotted closely to RB 39, which is resistant to WBD. Wild accessions genetically close to UF 273 type 1 include PA series, such as PA 169 and PA 4. PA 4 has resistance to WBD and BPR, and PA 169 is commonly used as a source of resistance to BPR and FPR (Turnbull and Hadley 2023). The accessions IMC 6 and IMC 31 are both characterized for having a low pod index (high yield) and resistance to Phytophthora (Turnbull and Hadley 2023) and are closely plotted to UF 273 type 1 in the PCoA (Fig. 2).
Agronomic traits showing divergence between wild and managed cacao
AMOVA analysis indicated that 5% of the total variation was caused by variation between wild and managed cacao, 37% by the variation among individuals, and 58% by the variation within individuals (Table 2). In addition, 11 SNPs showed FST values above 0.05, indicating moderate genetic differentiation between managed and wild cacao. Six of these SNPs had FST values greater than 0.10, suggesting strong divergence between the two groups at these markers (Table 3).
Three SNPs associated with yield traits showed high divergence between wild and cultivated cacao. Among these SNPs, Tcm004s00289192 (FST = 0.160, P < 0.001) was linked to dry bean weight and high yield, and TcSNP1370 (FST = 0.054, P < 0.001) was associated with the number of seeds per fruit. Tcm002s23708704 (FST = 0.107, P < 0.001) was related to the pod index, a measure of yield in commercial plantations. TcSNP1866 was associated with flower retention and had the highest FST = 0.260 (P < 0.001) between wild and managed cacao. Figure 4 shows the relative genotype frequency of SNPs showing pronounced divergence between wild and managed cacao and results of Fisher’s exact test.
Distribution of cacao genotypes for SNPs related to agronomic traits in the wild and managed cacao accessions. a SNP markers related to yield traits: flower retention (da Silva et al. 2016); dry bean weight/high yield and high pod index (Fernandes et al. 2020). b SNP markers related to disease resistance traits: witches’ broom disease (Royaert et al. 2016) and Ceratocystis wilt (Fernandes et al. 2018).*Significant difference between wild and managed cacao based on Fisher’s exact test (P < 0.05)
Managed and wild cacao showed also high divergence in disease resistance traits, especially in WBD resistance. We observed five SNPs significantly differentiated between managed and wild cacao that are associated with WBD resistance: Tcm006s19715703 (FST = 0.146, P < 0.001), Tcm004s00110232 (FST = 0.101, P < 0.001), TcSNP1230 (FST = 0.083, P < 0.001), Tcm003s33466269 (FST = 0.067, P < 0.001), and Tcm009s08066239 (FST = 0.063; P < 0.001). In addition, Tcm006s13222057 (FST = 0.155; P < 0.001) associated with resistance to CWC and Tcm009s40465466 (FST = 0.066; P < 0.001) related to FPR resistance were also significantly differentiated between managed and wild cacao (Table 3).
Figures 5 and 6 show the genotype frequencies for the SNPs Tcm004s00110232 and Tcm006s13222057 among cacao accessions resistant, tolerant, and susceptible to WBD and CW, respectively. Germplasm with resistance (n = 58) and tolerance (n = 25) to WBD showed a significantly higher frequency of the CT and TT genotypes (Tcm004s00110232), and the CC genotype showed a low frequency in resistant plants, while all susceptible plants (n = 9) showed the TT genotype (Fig. 5). In addition, germplasm evaluated as resistant (n = 28) and tolerant (n = 16) to CW showed a higher frequency of GG and GT (Tcm006s13222057) genotypes, while the TT genotype was common in susceptible (n = 21) plants. However, these differences were not statistically significant (P =0.7910, Fig. 6).
Genotype frequencies for the SNP Tcm004s00110232 related to witches’ broom disease (WBD) among cacao (Theobroma cacao L.) accessions with resistance, tolerance, or susceptibility to WBD. Number of accessions is provided in brackets. Reaction to WBD reported by the International Germplasm Cocoa Database (https://www.icgd.reading.ac.uk/index.php). *Significant difference between resistant-tolerant and susceptible groups based on Fisher’s exact test (P = 0.0026)
Genotype frequencies for the SNP Tcm006s13222057 associated with Ceratosystis wild disease among cacao (Theobroma cacao L.) accessions with resistance, tolerance, or susceptibility to Ceratosystis disease. Number of accessions is provided in parentheses. Reaction to Ceratosystis disease reported by the International Germplasm Cocoa Database (https://www.icgd.reading.ac.uk/index.php). No significant difference between resistant-tolerant and susceptible based on Fisher’s exact test (P = 0.7910)
Discussion
Low population structure in managed cacao
With our set of SNPs, we did not identify the ten genetic clusters observed earlier by Motamayor et al. (2008) based on 96 simple sequence repeat (SSR) markers. It is not surprising, considering a limited set of nonrandom 42 SNPs that we used in our study, which could be also under selection. However, we still identified four clusters reflecting the geographic origin of wild samples, with the second and third clusters being overrepresented in managed cacao, mainly consisting of hybrids (Fig. 1). This population pattern in managed cacao is in agreement with observations made by Cornejo et al. (2018). They analyzed the genome sequence of 200 cacao accessions and identified that cultivated cacao and man-made hybrids are mainly composed of two clusters, Criollo and Amelonado, with a low admixture of the Nacional cluster (Cornejo et al. 2018). Population genetic analysis demonstrated that Criollo was the first domesticated cacao and provides the foundations of cultivated cacao until today, mostly due to flavor and chocolate attributes (Lachenaud and Motamayor 2017; Motamayor et al. 2002). Our results confirm that a narrow cacao genetic diversity has been used in managed cacao likely due to retaining quality traits derived from Criollo observed in the second cluster in our analysis.
Cornejo et al. (2018) also observed that a higher Criollo ancestry in man-made hybrids is associated with low yield (seed productivity per year per plant) mainly due to the accumulation of deleterious mutations which led to reduced fitness in Criollo during the domestication process (Cornejo et al. 2018). The high contribution of Criollo to cultivated cacao helps to explain the low yield on average reported in cultivated cacao globally (Morrissey et al. 2019) and its susceptibility to BPR, WBD, and FP diseases (Ploetz 2016). To capture new genetic diversity and to broaden the genetic base for future breeding activities, new plant collections in northwestern Amazonia should be incorporated into cacao breeding programs (Nieves-Orduña et al. 2023).
In addition, the representation of the first and third clusters in managed cacao likely reflects gene introgression from the Pound Collection. The collection includes accessions MO, IMC, NA, SCA, and PA collected in the Peruvian Amazon while searching for trees resistant to WBD (Zhang et al. 2011). The Pound collection has been widely used in cacao breeding for developing disease resistance after the collapse of plantations in Surinam, Trinidad, Ecuador, and Brazil due to the introduction of WBD (Zhang et al. 2011). PA series also have been used for developing disease resistance against BPR and FPR (Zhang et al. 2011).
Genetic resources for cacao breeding
Guiana (GU) accessions were identified mainly in cluster four in our structure analysis and observed with a very low frequency in managed cacao (Fig. 1). The underutilization of Guiana accessions in cultivated cacao was also observed by Cornejo et al. (2018). These findings highlight opportunities to exploit the GU accessions for cacao breeding. Early studies reported cacao trees from French Guiana as novel sources of resistance to BPR and WBD and high yield (up to 1426 kg of dry seeds/year/ha) (Lachenaud et al. 2007; Paulin et al. 2008). This was supported by Ofori et al. (2020), who observed that GU accessions can broaden the genetic base of cacao breeding not only for BPR resistance but also for yield in Ghana (Ofori et al. 2020). In addition, Guiana accessions in Central America showed moderate resistance to FPR (Lachenaud et al. 2018), an essential agronomic trait for cacao cultivation in Tropical America (Evans et al. 1977; Gutiérrez et al. 2021b; Phillips-Mora et al. 2005, 2013). A detailed agronomic evaluation of multiple Guiana accessions is presented by Lachenaud et al. (2007). The best clones and those to be avoided were identified based on yield, disease resistance (BPR and WBD), and seed quality traits. The preselection of the best Guiana germplasm facilitates the introgression of new and valuable genetic diversity resources into cacao breeding programs (Lachenaud et al. 2007).
Yield and disease resistance: agronomic traits showing divergence
Cacao yield traits and disease resistance to WBD showed patterns of divergence between wild and managed cacao accessions (Table 3). These patterns are associated with the history of cacao domestication, cultivation, and selection. After initial selection for pulp flavor and seed traits in the sister Curaray population on the Ecuador-Colombia border, human selection led to the domestication of Criollo cacao ~ 3600 years ago (Clement et al. 2010; Cornejo et al. 2018). In addition, evidence based on ancient cacao DNA supports the consumption of cacao in the Ecuadorian Amazon around 5300 years ago; the DNA analyzed was closer to the Curaray and Purus clusters than to other cacao clusters (Zarrillo et al. 2018). The genetic cost of cacao domestication led to the accumulation of deleterious mutations, susceptibility to diseases, and low yield in Criollo (Cornejo et al. 2018). Criollo with larger seed size, white cotyledons, and reduced bitterness was distributed and cultivated outside the Upper Amazon by native Americans in Northern Colombia and Mesoamerica (Cornejo et al. 2018; Motamayor et al. 2002).
Criollo cultivation expanded during colonial times through Tropical America (Bartley 2005), especially in Trinidad, where some blast destroyed the crop in 1727 (Díaz-Valderrama et al. 2020). This collapse in cacao production led to the introduction of new plant material from upper Amazonia, which hybridized naturally with the cultivated Criollo and formed a hybrid cultivated cacao known as Trinitario germplasm (Zhang et al. 2011). From these vigorous hybrids, a breeding program was started in 1930 by the Imperial College of Tropical Agriculture of Trinidad focused on yield traits (Toxopeus 1969). The best trees were selected based on the number of seeds per pod and bean weight, which resulted in 100 trees known as the Imperial College Selection (Toxopeus 1969). The mean seed weight was an important trait due to the premium price for large seeds in the market (Toxopeus 1969), a trait that still is considered of economic importance in modern cacao breeding programs (Bekele et al. 2022). In addition, after the introduction and impact of WBD in 1932, new wild material with disease-resistance traits was needed in Trinidad. The material searched and collected from the Peruvian Amazon in 1937–1938 includes accessions known as the Pound Collection. This collection created the genetic base for developing breeding resistance against WBD globally (Bartley 2005; Díaz-Valderrama et al. 2020; Evans 2016; Zhang et al. 2011).
In Brazil, the low genetic diversity of cultivated cacao led to the collapse of cacao economies in 1989 due to the introduction of WBD in Bahia (Bennett 2003; Evans 2016). In response, breeding programs started in Brazil to broaden the genetic base of cultivated cacao and evaluated germplasm collections for developing resistance against WBD (Bennett 2003).
SNPs related to flower setting (sexual compatibility)
Within accessions analyzed here, we observed TcSNP1866 with the highest FST value of 0.260 showing pronounced divergence between wild and managed cacao (Table 3). This SNP was identified in a GWAS study using 295 trees and 5301 SNPs, and incompatibility was measured as the frequency of flower retention 15 days after self-pollination (on average, 21 flowers were self-pollinated per tree) instead of a yes/no trait (da Silva et al. 2016). Previous studies in cacao highlighted that thousands of individuals are necessary to avoid bias in the estimation of SNP effects associated with small sample sizes (da Silva et al. 2016). In addition, flower dropping is influenced by rainfall, high temperature, or insects’ attack which can introduce underestimation in flower retention values (da Silva et al. 2016). Thus, the effect of genotype CC (TcSNP1866) associated with a high percentage (33%) of flower retention may be influenced by small sampling sizes and/or environmentally induced flower dropping (da Silva et al. 2016). From the breeders’ perspective, to select self-compatible trees, it is recommended to implement genomic selection which considers thousands of SNPs (da Silva et al. 2016).
Clones common to cacao breeding, including disease-resistant and commercial trees are self-incompatible (López et al. 2021; Phillips-Mora et al. 2013). Self-incompatibility in cacao requires plantation designs where cross-compatible clones are established to foster the exchange of pollen and field production (López et al. 2021; Phillips-Mora et al. 2013). Since incompatibility is a limiting factor in cacao yield, a common breeding objective is to avoid self-incompatible trees in breeding populations (López et al. 2021).
Although we do not have information on the percentage of flower retention after self-pollinations in the accessions genotyped here that would allow us to validate the phenotypes associated with TcSNP1866, we observed a significantly lower frequency of the genotype CC associated with self-compatibility (da Silva et al. 2016) in managed cacao (Fig. 4A), which may reflect the fact that breeding populations and advanced selections include self-incompatible trees. For example, self-compatibility was evaluated in commercial clones such as EET (62, 95, 96, 400), CAUCASIA (37, 39, 43, 47), ICS (1, 6, 39, 60, 95), and UF (29, 273, 613, 667, 676) (López et al. 2021). These clones (5 to 10 years old) were self-pollinated, and the mean fruit set was 24% across clones, indicating partial self-incompatibility (López et al. 2021). The genotypes (n = 18) for these incompatible clones indicated a higher proportion of the homozygous GG (56%), followed by CC (39%) and CG (6%).
As self-compatibility is not absolute in cacao (Lopes et al. 2022), the observation of the genotype CC (TcSNP1866) at higher frequency in wild cacao reflects some degree of self-compatibility in wild populations (Fig. 4A). This could be due to the geographic origin of wild trees observed isolated along river basins and pollinated by midges with a reduced range of movement (Lopes et al. 2022). Accordingly, levels of homozygosity above 70% were observed among wild accessions genotyped with genome-wide SNPs (3 K) (Lopes et al. 2022).
SNPs related to yield and pod index
Cacao yield (kg of dry seeds/year/ha) is a polygenic complex trait, at least 40 candidate genes of functional importance encode embryo and seed development, protein synthesis, carbohydrate transport, and lipid biosynthesis and transport (Bekele et al. 2022). In addition, yield components such as the number of pods produced per tree and bean dry weight per pod are influenced by genotypes, environment, agronomic management (e.g., fertilization, shade, irrigation), and diseases (Bartley 2005; Doaré et al. 2020; Fernandes et al. 2020; Phillips-Mora et al. 2013; Solís Bonilla et al. 2022). Disease pressure also influences yield, as high-yielding clones with disease resistance traits exhibited a low disease incidence (Phillips-Mora et al. 2013). But, the same clone can exhibit varying yield potential due to exposition to different pathogen strains (variations in disease pressure) (Jaimes et al. 2011, 2019). Disease management in cacao farms also affects the yield potential of clones (Jaimez et al. 2020). For example, removal of FPR-infected pods is a common practice to reduce disease incidence and increase tree productivity (Jaimes et al. 2019; Jaimez et al. 2020).
The SNP Tcm004s00289192 related to yield showed a pronounced divergence between wild and managed cacao (Table 3). Within accessions analyzed here, we observed the homozygous genotype GG more frequently in managed cacao (Fig. 4A). Among the accessions with the GG genotype are advanced selections such as ICS, UF, TARS, and EET clones. The heterozygous genotype AG was observed in high-yielding clones such as TARS, CATIE R6, CCN 51, and VB clones. The allele G (Tcm004s00289192) is associated with a higher yield (Fernandes et al. 2020). Fernandes et al. (2020) identified copy-number variations of SWEET (Sugar Will Eventually be Exported Transporters) genes between the markers Tcm004s00289192 and Tcm004s00615809 on chromosome IV. Likewise, a recent GWAS using a diverse cacao germplasm in Trinidad identified SNP markers within the genes SWEET17 (chromosome 4) and SWEET2 (chromosome 7) related to cacao yield traits such as pod index and seed number (Bekele et al. 2022). Sweet proteins are a family of sugar transporters important for plants' biological processes such as growth, development, and response to abiotic and biotic stresses (Singh et al. 2023). SWEET genes in cultivated tree species such as apple (Malus x domestica) contribute to fruit sugar accumulation (Zhen et al. 2018), and in Litchi chinensis, they played roles in fruit development, growth, and seed development (Xie et al. 2019). SWEET4 also contributed to rice and maize domestication by enhancing seed sizes as it facilitates sugar transport during grain filling (Sosso et al. 2015). Further characterization of cacao SWEET genes in wild and cultivated cacao can help to identify alleles contributing to cacao domestication and validate useful alleles for improving yield traits in cacao.
In cacao breeding, pod index is defined as the number of pods (fruits) necessary to produce 1 kg of dry seeds; a low pod index (14–20) is associated with high yield potential and heavier seeds (Bekele et al. 2020; Fernandes et al. 2020). Early breeding programs in Trinidad (1940s) focused on yield and pod index; selected trees exhibited a low pod index (18) and higher yield (1000 kg/ha) (Toxopeus 1969). The pod index also showed a high narrow-sense heritability (0.64) and stability across different sites at two farms in Bahia, Brazil, making the trait a target of selection (DuVal et al. 2017). In addition, a low pod index is also selected in germplasm collections and breeding populations because less healthy fruits are necessary to produce 1 kg of dry cacao, meaning less costs associated with harvesting and pod breaking (Bekele et al. 2020; Solís Bonilla et al. 2022). The SNP Tcm002s23708704 related to pod index showed a divergence between wild and managed cacao (Table 3), and Fernandes et al. (2020) reported the A allele to be associated with high pod index. We observed the genotype AA (SNP Tcm002s23708704) at low frequency in managed cacao compared to wild cacao (Fig. 4A). Although we do not have phenotypic information for the accession genotyped here, the low frequency of the A allele in managed cacao may reflect the continued selection favoring a low pod index in breeding populations and cultivated cacao.
SNPs related to witches’ broom disease (WBD) resistance
WBD is a devastating disease for cacao cultivation (Evans 2016). To improve disease resistance, cacao breeders use SCA 6 and SCA 12 clones as primary sources for breeding WBD resistance (Gutiérrez et al. 2016). However, due to the susceptibility of SCA clones under high disease pressure, there was a need to identify new sources of resistance. In this effort, Pereira et al. (2021) identified new clones, such as C SUL-3 and GU-171, resistant to WBD in Bahia, Brazil. In addition, new expeditions to the Peruvian Amazon collected 280 cacao trees to diversify the gene pool and resistance to WBD (Durham 2011). Native to Amazonia but widely distributed in South America, WBD has different pathogen strains (Lisboa et al. 2020; Ploetz 2016). Thus, a major breeding objective is to develop cultivars with broad WBD disease resistance (Meinhardt et al. 2008). WBD resistance is a complex trait involving at least sixteen candidate resistance genes identified using genome-wide association studies (GWAS) (Osorio-Guarín et al. 2020) and quantitative trait locus (QTL) analysis (Chia Wong et al. 2022; Mournet et al. 2020; Royaert et al. 2016).
The SNP Tcm004s00110232 related to WBD resistance showed divergence between wild and managed cacao (Table 3). We inferred the data on reaction to WBD for 92 samples (30%) from the ICGD (Turnbull and Hadley 2023), while 219 (70%) have no data available. The phenotypic data reported are from different WBD studies (Turnbull and Hadley 2023), and due to region-specific differences among studies, likely responses to different pathogen strains were assessed. The genotypes CT and TT (Tcm004s00110232) are highly represented in resistant (n = 58) and tolerant (n = 25) plants (Fig. 5). The genotype CT observed in the clone TSH 1188 was reported as WBD-resistant by Royaert et al. (2016). In addition, the genotype CC (Tcm004s00110232) was observed only in resistant germplasm (Fig. 5). These results are in agreement with Lachenaud et al. (2007) that reported the observed homozygous CC in resistant to WBD Guiana clones (GU 171 /C; GU 219 /F; GU 221 /C; GU 261 /P; GU 277 /G) (Lachenaud et al. 2007; Turnbull and Hadley 2023).
The genotype TT (Tcm004s00110232) was detected in all nine accessions reported as susceptible to WBD (Fig. 5), but more samples for this category with other genotypes could probably be detected. This reaction to WBD is explained by distinct variants of M. perniciosa (range of pathogen aggressiveness) and pathogen adaptation to resistant trees such as SCA 6 (Artero et al. 2017; Pereira et al. 2021; Royaert et al. 2016). For example, in Bahia, Brazil, a decrease in resistance of SCA clones to WBD has been reported due to continuous and high disease pressure (Pereira et al. 2021). The fact that the SCA clones have been the main source for breeding WBD resistance motivated the search and evaluation of new sources of disease resistance from different geographical areas (Durham 2011; Pereira et al. 2021). In addition, disease resistance is a polygenic trait with each gene explaining a relatively small portion of the variation in disease resistance.
The majority of wild accessions here genotyped correspond to a subset of samples collected during the 1930–1940s in Peru by Frederick J. Pound (see “Materials and methods” section), whose main purpose was collecting cacao trees exhibiting WBD disease resistance traits (Bartley 2005; Zhang et al. 2011). This explains why the favorable allele T (Tcm004s00110232) was observed at a high frequency in wild cacao (Fig. 4B). The high frequency of genotype CT in managed cacao reflects the hybridization of managed cacao with wild cacao accessions. For example, a total of 191 crosses used SCA 6 (low yield but resistant to WBD) as a parent for the incorporation of disease-resistance traits in cultivated cacao (Turnbull and Hadley 2023). As a result, commercial hybrids such as ICS, TSH, EET, EQX, and TARS with various levels of WBD resistance were developed using SCA 6 (Turnbull and Hadley 2023).
SNPs related to Ceratocystis wilt (CW) resistance
CW targets the cacao vascular system and causes the death of infected trees (Engelbrecht et al. 2007). The disease is caused by the host-specialized fungus Ceratocystis cacaofunesta, which is native to South America (Western Ecuador and Southwest Brazil) (Engelbrecht et al. 2007). The disease is geographically restricted to Tropical America. Still, it threatens the cacao economy because it can be dispersed to important cacao-producing regions such as West Africa and Asia (Engelbrecht et al. 2007). Early reports in the 1950s described CW causing damage to cocoa farms in Colombia, Ecuador, Costa Rica, and Trinidad, and in 1997, it was observed in Bahia, Brazil (Cabrera et al. 2016). Breeding for disease resistance is gaining attention as germplasm selected for WBD resistance in Brazil such as “Theobahia” shows susceptibility to CW (Fernandes et al. 2018; Lopes et al. 2011).
The SNP Tcm006s13222057 is associated with resistance to CW (Fernandes et al. 2018). This SNP shows significant differences between wild and cultivated cacao populations (Table 3). To investigate the relationship between SNP genotypes and CW resistance, we searched in the ICGD for the reaction to CW among the accessions genotyped (Turnbull and Hadley 2023). Only 65 accessions (21%) have reported data on the CW reaction (resistant, tolerant, or susceptible), while 249 accessions (79%) have no data available. Among the accessions with reported data, we observed that the resistant (n = 28) and tolerant (n = 16) groups had a high frequency of the G allele (Tcm006s13222057). The genotypes GT and GG were more frequent in the resistant and tolerant group, respectively, while in the susceptible group, the genotype TT is prevalent (Fig. 6). However, these differences were not significant, likely, due to small sample size. These results are, however, consistent with the QTL analysis by Fernandes et al. (2018) that identified the G allele as a marker for CW resistance.
We observed the genotype GG at high frequency in wild cacao (Fig. 4B). Among these accessions, the IMC clones (IMC 11, 31, 47, 60, and 67), PA 121, POUND (12, 12A), SCA (6, 12), and U (26, 70) were reported as resistant to CW (Turnbull and Hadley 2023). The wild accession IMC 67 is frequently used in cacao breeding programs for its resistance to CW and vegetative vigor and is widely used as a rootstock (Cabrera et al. 2016; Osorio Montoya et al. 2022). IMC 67 was used in 145 crosses for developing cultivars and breeding populations of the series CEPEC, EET, EQX, TSH, and TSH (Turnbull and Hadley 2023). However, IMC 67 was reported to be resistant to the Ecuadorian isolate but susceptible to the Brazilian strain (Cabrera et al. 2016), highlighting the need for new (wild) sources against different CW strains. In addition, we observed the heterozygous genotype GT at high frequency in managed cacao (Fig. 4B). Heterozygous clones reported as disease resistant to CW include EET clones (399, 400), SC 20, ICS clones (6, 40, 95), TSH (1188, 595), UF (29, 650), and VB 650 and VB 681, which is recommended for large scale planting in Bahia, Brazil (Lopes et al. 2011; Turnbull and Hadley 2023).
Conclusions and research questions
Accession structure of wild and managed cacao
Our results confirm a narrow genetic diversity in managed cacao likely reflecting Criollo ancestry of accessions traditionally used to select for chocolate quality. Among the germplasm analyzed, managed cacao (cultivated and breeding populations) showed introgression of wild cacao collected in western Amazonia (Peru, Ecuador), but much less contribution of Guiana clones. New collection trips should be done to broaden the genetic base for cultivated cacao to improve agronomic traits, such as urgently needed FPR disease resistance. Potential areas for cacao germplasm collection are highlighted by Nieves-Orduña et al. (2023).
New genetic resources for cacao breeding
Guiana accessions represent a genetic resource underutilized in managed cacao. Agronomic evaluations of this germplasm group showed favorable yield traits (e.g., accession GU 285) and resistance against WBD and BPR (Lachenaud et al. 2007). Cacao breeding programs can obtain Guiana clones free of pathogens through the ICQC and use them in perspective breeding experiments to exploit their potential under a hybridization scheme. Developing of cacao inbred lines can support hybrid breeding, but homozygosity and genetic distance between accessions should be considered to maximize heterosis (Akpertey et al. 2022; Lopes et al. 2022).
Validation of SNPs and cacao breeding
Cacao SNPs associated with disease resistance and yield traits should be validated across diverge germplasm added with standard phenotypic information obtained from multi-environment studies and using trees with the same age as well as clonal replicates. This validation facilitates the identification of useful major QTL for cultivar development by different breeding programs. In addition, breeding programs can adopt and optimize genomic selection. Previous studies have demonstrated to improve selection if individual effects of multiple SNP markers are considered in complex cacao traits such as yield and disease resistance (Bekele et al. 2022; McElroy et al. 2018; Romero-Navarro et al. 2017).
Candidate genes of cacao domestication
Disease resistance and yield traits showed divergence between wild and managed cacao, probably reflecting selection during domestication, cultivation, and breeding efforts. Further analysis with a diverse natural cacao population, cultivars (e.g., CEPEC´s germplasm of Brazil), and clones developed by chocolate companies for specific farm conditions, using high-density SNP genotyping panels (cacao 15K SNP array) will help identify signatures of selection in candidate genes with high phenotypic impact on agronomic traits. In addition, previous studies with SSR markers revealed a low number of private alleles in the traditional cultivars (Criollo, Amelonado, and Nacional) when compared to the wild Amazonian cacao groups likely due to bottlenecks during the selection and domestication process (Motamayor et al. 2008; Clement et al. 2010). New analyses with more SNP markers are needed to confirm this pattern in the distribution of private alleles among different cacao germplasm groups.
Other traits of economic importance
Plant architecture is a trait of economic importance not widely investigated in breeding programs to improve harvest index (ratio of seeds to total below biomass produced). The breeding challenge is to reduce plant height to facilitate harvesting, but without affecting yield. Mustiga et al. (2018) proposed selecting trees with small trunk diameter, lower branch angles, and high yield to increase plant density. In addition, market restrictions in cacao-based products with high cadmium levels have led to evaluating germplasm and identifying valuable genetic sources to introduce low cadmium traits into commercial clones (Lewis et al. 2018).
Cacao cultivation requires effective climate adaptation, particularly in Brazil and West Africa (Araújo et al. 2024; Dzandu et al. 2021; Schroth et al. 2016). Root traits (such as angle and biomass) that enhance water use efficiency should be characterized in different cacao genotypes (Lahive et al. 2019). For example, among 18 genotypes in an irrigated and non-irrigated experiment, the commercial clone CNN 51 showed a better physiological response to water stress (Araújo et al. 2024). In addition, cacao trees growing (with precipitations ≤ 1000 mm/year) in the forests of Peru and Ecuador should be exploited for building breeding populations with drought tolerance traits (Nieves-Orduña et al. 2023).
References
Agena Bioscience (2015) Assay Design Suite v2.0: User Guide. USG-CUS-040 Rev02
Agena Bioscience (2019a) iPLEX Pro and Gold Reagents: User Guide. USG-CUS-071 Rev03
Agena Bioscience (2019b) MassARRAY Typer: User Guide. USG-CUS-027 Rev05
Akpertey A, Padi FK, Meinhardt L, Zhang D (2022) Relationship between genetic distance based on single nucleotide polymorphism markers and hybrid performance in Robusta coffee (Coffea canephora). Plant Breed 141:286–300. https://doi.org/10.1111/pbr.13005
Allen JB (1988) Geographical variation and population biology in wild Theobroma cacao. The University of Edinburgh, PhD
Araújo MS, Chaves SF, Pereira GR, Guimarães MH, Alves AK, Dias LA, Souza CA, Aguilar MA (2024) Multi-trait selection for nutritional and physiological quality of cacao genotypes in irrigated and non-irrigated environments. Sci Rep 14:6368. https://doi.org/10.1038/s41598-024-56556-7
Artero AS, Silva JQ, Albuquerque PSB, Bressan EA, Leal GA, Sebbenn AM, Griffith GW, Figueira A (2017) Spatial genetic structure and dispersal of the cacao pathogen Moniliophthora perniciosa in the Brazilian Amazon. Plant Pathol 66:912–923. https://doi.org/10.1111/ppa.12644
Barlow J, Gardner TA, Lees AC, Parry L, Peres CA (2012) How pristine are tropical forests? An ecological perspective on the pre-Columbian human footprint in Amazonia and implications for contemporary conservation. Biol Conserv 151:45–49. https://doi.org/10.1016/j.biocon.2011.10.013
Bartley BGD (2005) The genetic diversity of cacao and its utilization. CABI Pub, Cambridge MA
Bekele F, Phillips-Mora W (2019) Cacao (Theobroma cacao L.) breeding. In: Al-Khayri JM, Jain SM, Johnson DV (eds) Advances in plant breeding strategies: industrial and food crops, vol 46. Springer International Publishing, Cham, pp 409–487
Bekele F, Bidaisee G, Singh H, Saravanakumar D (2020) Morphological characterisation and evaluation of cacao (Theobroma cacao L.) in Trinidad to facilitate utilisation of Trinitario cacao globally. Genet Resour Crop Evol 67:621–643. https://doi.org/10.1007/s10722-019-00793-7
Bekele F, Bidaisee G, Allegre M, Argout X, Fouet O, Boccara M, Saravanakumar D, Bekele I, Lanaud C (2022) Genome-wide association studies and genomic selection assays made in a large sample of cacao (Theobroma cacao L.) germplasm reveal significant marker-trait associations and good predictive value for improving yield potential. PLoS ONE 17:e0260907. https://doi.org/10.1371/journal.pone.0260907
Bennett AB (2003) Out of the Amazon: Theobroma cacao enters the genomic era. Trends Plant Sci 8:561–563. https://doi.org/10.1016/j.tplants.2003.10.004
Boza EJ, Motamayor JC, Amores FM, Cedeño-Amador S, Tondo CL, Livingstone DS, Schnell RJ, Gutiérrez OA (2014) Genetic characterization of the cacao cultivar CCN 51: its impact and significance on global cacao improvement and production. J Am Soc Horti Sci 139:219–229. https://doi.org/10.21273/JASHS.139.2.219
Cabrera OG, Molano EPL, José J, Álvarez JC, Pereira GAG (2016) Ceratocystis wilt pathogens: history and biology—highlighting C. cacaofunesta, the causal agent of Wilt Disease of cacao. In: Bailey BA, Meinhardt LW (eds) Cacao Diseases. Springer International Publishing, Cham, pp 383–428
CacaoNet (2012) A global strategy for the conservation and use of cacao genetic resources, as the foundation for a sustainable cocoa economy. Bioversity International, Montpellier, France
Chia Wong JA, Clement DPL, Mournet P, dos Santos Nascimento A, Solís Bonilla JL, Lopes UV, Pires JL, Gramacho KP (2022) A high-density genetic map from a cacao F2 progeny and QTL detection for resistance to witches’ broom disease. Tree Genet Genomes 18:31. https://doi.org/10.1007/s11295-022-01562-x
Clement C, de Cristo-Araújo M, Coppens D’Eeckenbrugge G, Alves Pereira A, Picanço-Rodrigues D (2010) Origin and domestication of native Amazonian crops. Diversity 2:72–106. https://doi.org/10.3390/d2010072
Clement CR, Denevan WM, Heckenberger MJ, Junqueira AB, Neves EG, Teixeira WG, Woods WI (2015) The domestication of Amazonia before European conquest. Proc R Soc Lond B 282:20150813. https://doi.org/10.1098/rspb.2015.0813
Coe SD, Coe MD (2013) The true history of chocolate. Thames & Hudson, London
Cornejo OE, Yee M-C, Dominguez V, Andrews M, Sockell A, Strandberg E, Livingstone D, Stack C, Romero A, Umaharan P, Royaert S, Tawari NR, Ng P, Gutierrez O, Phillips W, Mockaitis K, Bustamante CD, Motamayor JC (2018) Population genomic analyses of the chocolate tree, Theobroma cacao L., provide insights into its domestication process. Commun Biol 1:167. https://doi.org/10.1038/s42003-018-0168-6
da Silva MR, Clément D, Gramacho KP, Monteiro WR, Argout X, Lanaud C, Lopes U (2016) Genome-wide association mapping of sexual incompatibility genes in cacao (Theobroma cacao L.). Tree Genet Genomes 12:15. https://doi.org/10.1007/s11295-016-1012-0
Dand R (2011) The international cocoa trade. Elsevier, Cambridge UK
Daymond A (2018) Safe handling and movement of cocoa germplasm for breeding. In: Umaharan P (ed) Achieving sustainable cultivation of cocoa. Burleigh Dodds Science Publishing, Cambridge UK, pp 101–110. https://doi.org/10.19103/AS.2017.0021.05
Díaz-Valderrama JR, Leiva-Espinoza ST, Aime MC (2020) The history of cacao and its diseases in the Americas. Phytopathology 110:1604–1619. https://doi.org/10.1094/PHYTO-05-20-0178-RVW
Doaré F, Ribeyre F, Cilas C (2020) Genetic and environmental links between traits of cocoa beans and pods clarify the phenotyping processes to be implemented. Sci Rep 10:9888. https://doi.org/10.1038/s41598-020-66969-9
Durham S (2011) Peruvian cacao collections trip yields treasures. Agric Res 59:8–10. https://agresearchmag.ars.usda.gov/2011/sep/cacao
DuVal A, Gezan SA, Mustiga G, Stack C, Marelli J-P, Chaparro J, Livingstone D, Royaert S, Motamayor JC (2017) Genetic parameters and the impact of off-types for Theobroma cacao L. in a breeding program in Brazil. Front Plant Sci 8:2059. https://doi.org/10.3389/fpls.2017.02059
Dzandu E, Enu-Kwesi L, Markwei CM, Ayeh KO (2021) Screening for drought tolerance potential of nine cocoa (Theobroma cacao L.) genotypes from Ghana. Heliyon 7:e08389. https://doi.org/10.1016/j.heliyon.2021.e08389
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Engelbrecht CJ, Harrington TC, Alfenas A (2007) Ceratocystis wilt of cacao-a disease of increasing importance. Phytopathology 97:1648–1649. https://doi.org/10.1094/PHYTO-97-12-1648
Evans HC (2016) Witches’ broom disease (Moniliophthora perniciosa): history and biology. In: Bailey BA, Meinhardt LW (eds) Cacao Diseases. Springer International Publishing, Cham, pp 137–177
Evans HC, Edwards DF, Rodríguez M (1977) Research on cocoa diseases in Ecuador: past and present. PANS 23:68–80. https://doi.org/10.1080/09670877709412399
Fernandes LDS, Royaert S, Corrêa FM, Mustiga GM, Marelli J-P, Corrêa RX, Motamayor JC (2018) Mapping of a major QTL for Ceratocystis Wilt disease in an F1 population of Theobroma cacao. Front Plant Sci 9:155. https://doi.org/10.3389/fpls.2018.00155
Fernandes LDS, Correa FM, Ingram KT, Almeida A-A, Royaert S (2020) QTL mapping and identification of SNP-haplotypes affecting yield components of Theobroma cacao L. Hortic Res 7:26. https://doi.org/10.1038/s41438-020-0250-3
Fick SE, Hijmans RJ (2017) WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol 37:4302–4315. https://doi.org/10.1002/joc.5086
Gutiérrez O, Campbell A, Phillips-Mora W (2016) Breeding for disease resistance in cacao. In: Bailey BA, Meinhardt LW (eds) Cacao Diseases. Springer International Publishing, Cham, pp 567–609
Gutiérrez O, Martinez K, Zhang D, Livingstone D, Turnbull CJ, Motamayor JC (2021a) Selecting SNP markers reflecting population origin for cacao (Theobroma cacao L.) germplasm identification. Beverage Plant Res 1:1–9. https://doi.org/10.48130/BPR-2021-0015
Gutiérrez O, Puig A, Phillips-Mora W, Bailey BA, Ali SS, Mockaitis K, Schnell RJ, Livingstone D, Mustiga G, Royaert S, Motamayor JC (2021b) SNP markers associated with resistance to frosty pod and black pod rot diseases in an F1 population of Theobroma cacao L. Tree Genet Genomes 17:106. https://doi.org/10.1007/s11295-021-01507-w
Hoang NT, Kanemoto K (2021) Mapping the deforestation footprint of nations reveals growing threat to tropical forests. Nat Ecol Evol 5:845–853. https://doi.org/10.1038/s41559-021-01417-z
Jaimes Y, Aránzazu F, Rodríguez E, Martínez N (2011) Behavior of introduced regional clones of Theobroma cacao toward the infection Moniliophthora roreri in three different regions of Colombia. Agron Colomb 29:361–371. http://www.scielo.org.co/scielo.php?script=sci_arttext&pid=S0120-99652011000200002
Jaimes Y, Ribeyre F, Gonzalez C, Rojas J, Furtado E, Cilas C (2019) Factors affecting the dynamics of Frosty Pod Rot in the main cocoa areas of Santander State, Colombia. Plant Dis 103:1665–1673. https://doi.org/10.1094/PDIS-10-18-1761-RE
Jaimez R, Vera D, Mora A, Loor R, Bailey B (2020) A disease and production index (DPI) for selection of cacao (Theobroma cacao) clones highly productive and tolerant to pod rot diseases. Plant Pathol 69:698–712. https://doi.org/10.1111/ppa.13156
Kalischek N, Lang N, Renier C, Daudt RC, Addoah T, Thompson W, Blaser-Hart WJ, Garrett R, Schindler K, Wegner JD (2023) Cocoa plantations are associated with deforestation in Côte d’Ivoire and Ghana. Nat Food 4:384–393. https://doi.org/10.1038/s43016-023-00751-8
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191. https://doi.org/10.1111/1755-0998.12387
Lachenaud P (1997) Genetic/taxonomic structuring of the Theobroma cacao L. species fresh hypotheses. INGENIC Newsl 3:10–11
Lachenaud P (2015) Wild cacao trees (Theobroma cacao L.) in French Guiana. Acad J Suriname 6:555–560
Lachenaud P, Motamayor JC (2017) The Criollo cacao tree (Theobroma cacao L.): a review. Genet Resour Crop Evol 64:1807–1820. https://doi.org/10.1007/s10722-017-0563-8
Lachenaud P, Paulin D, Ducamp M, Thevenin J-M (2007) Twenty years of agronomic evaluation of wild cocoa trees (Theobroma cacao L.) from French Guiana. Sci Hortic 113:313–321. https://doi.org/10.1016/j.scienta.2007.05.016
Lachenaud P, Clément D, Argout X, Scalabrin S, Doaré F (2018) The Guiana cacao genetic group (Theobroma cacao L.): a new core collection in French Guiana. Bot Lett 165:248–254. https://doi.org/10.1080/23818107.2018.1465466
Lahive F, Hadley P, Daymond AJ (2019) The physiological responses of cacao to the environment and the implications for climate change resilience. A review. Agron Sustain Dev 39:131. https://doi.org/10.1007/s13593-018-0552-0
Lanaud C, Vignes H, Utge J, Valette G, Rhoné B, Garcia Caputi M, Angarita Nieto NS, Fouet O, Gaikwad N, Zarrillo S, Powis TG, Cyphers A, Valdez F, Olivera Nunez SQ, Speller C, Blake M, Valdez F, Raymond S, Rowe SM, Duke GS, Romano FE, Loor Solórzano RG, Argout X (2024) A revisited history of cacao domestication in pre-Columbian times revealed by archaeogenomic approaches. Sci Rep 14:2972. https://doi.org/10.1038/s41598-024-53010-6
Langella O (2001) Populations, 1.2.31: Population genetic software. Free Software Foundation. https://bioinformatics.org/populations/
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. https://doi.org/10.1093/nar/gkab301
Levis C, Costa FRC, Bongers F, Peña-Claros M, Clement CR, Junqueira AB, Neves EG, Tamanaha EK, Figueiredo FOG, Salomão RP, Castilho CV, Magnusson WE, Phillips OL, Guevara JE, Sabatier D, Molino J-F, López DC, Mendoza AM, Pitman NCA, Duque A, Vargas PN, Zartman CE, Vasquez R, Andrade A, Camargo JL, Feldpausch TR, Laurance SGW, Laurance WF, Killeen TJ, Nascimento HEM, Montero JC, Mostacedo B, Amaral IL, Guimarães Vieira IC, Brienen R, Castellanos H, Terborgh J, Carim MdJV, Guimarães JRdS, Coelho LdS, Matos FDdA, Wittmann F, Mogollón HF, Damasco G, Dávila N, García-Villacorta R, Coronado ENH, Emilio T, Filho DdAL, Schietti J, Souza P, Targhetta N, Comiskey JA, Marimon BS, Marimon B-H, Neill D, Alonso A, Arroyo L, Carvalho FA, de Souza FC, Dallmeier F, Pansonato MP, Duivenvoorden JF, Fine PVA, Stevenson PR, Araujo-Murakami A, Aymard CGA, Baraloto C, do Amaral DD, Engel J, Henkel TW, Maas P, Petronelli P, Revilla JDC, Stropp J, Daly D, Gribel R, Paredes MR, Silveira M, Thomas-Caesar R, Baker TR, da Silva NF, Ferreira LV, Peres CA, Silman MR, Cerón C, Valverde FC, Di Fiore A, Jimenez EM, Mora MCP, Toledo M, Barbosa EM, Bonates LCdM, Arboleda NC, Farias EdS, Fuentes A, Guillaumet J-L, Jørgensen PM, Malhi Y, de Andrade-Miranda IP, Phillips JF, Prieto A, Rudas A, Ruschel AR, Silva N, von Hildebrand P, Vos VA, Zent EL, Zent S, Cintra BBL, Nascimento MT, Oliveira AA, Ramirez-Angulo H, Ramos JF, Rivas G, Schöngart J, Sierra R, Tirado M, van der Heijden G, Torre EV, Wang O, Young KR, Baider C, Cano A, Farfan-Rios W, Ferreira C, Hoffman B, Mendoza C, Mesones I, Torres-Lezama A, Medina MNU, van Andel TR, Villarroel D, Zagt R, Alexiades MN, Balslev H, Garcia-Cabrera K, Gonzales T, Hernandez L, Huamantupa-Chuquimaco I, Manzatto AG, Milliken W, Cuenca WP, Pansini S, Pauletto D, Arevalo FR, Reis NFC, Sampaio AF, Giraldo LEU, Sandoval EHV, Gamarra LV, Vela CIA, Steege H (2017) Persistent effects of pre-Columbian plant domestication on Amazonian forest composition. Science 355:925–931. https://doi.org/10.1126/science.aal0157
Lewis C, Lennon AM, Eudoxie G, Umaharan P (2018) Genetic variation in bioaccumulation and partitioning of cadmium in Theobroma cacao L. Sci Total Environ 640–641:696–703. https://doi.org/10.1016/j.scitotenv.2018.05.365
Lisboa DO, Evans HC, Araújo JPM, Elias SG, Barreto RW (2020) Moniliophthora perniciosa, the mushroom causing witches’ broom disease of cacao: insights into its taxonomy, ecology and host range in Brazil. Fungal Biol 124:983–1003. https://doi.org/10.1016/j.funbio.2020.09.001
Lopes UV, Monteiro WR, Pires JL, Clement D, Yamada MM, Gramacho KP (2011) Cacao breeding in Bahia, Brazil: strategies and results. Theor Appl Genet 11:73–81. https://doi.org/10.1590/S1984-70332011000500011
Lopes UV, Pires JL, Gramacho KP, Grattapaglia D (2022) Genome-wide SNP genotyping as a simple and practical tool to accelerate the development of inbred lines in outbred tree species: an example in cacao (Theobroma cacao L.). PLoS ONE 17:e0270437. https://doi.org/10.1371/journal.pone.0270437
López ME, Ramírez OA, Dubón A, Ribeiro THC, Díaz FJ, Chalfun-Junior A (2021) Sexual compatibility in cacao clones drives arrangements in the field leading to high yield. Sci Hortic 287:110276. https://doi.org/10.1016/j.scienta.2021.110276
McElroy MS, Navarro AJR, Mustiga G, Stack C, Gezan S, Peña G, Sarabia W, Saquicela D, Sotomayor I, Douglas GM, Migicovsky Z, Amores F, Tarqui O, Myles S, Motamayor JC (2018) Prediction of cacao (Theobroma cacao) resistance to Moniliophthora spp. diseases via genome-wide association analysis and genomic selection. Front Plant Sci 9:343. https://doi.org/10.3389/fpls.2018.00343
Meinhardt LW, Rincones J, Bailey BA, Aime MC, Griffith GW, Zhang D, Pereira GAG (2008) Moniliophthora perniciosa, the causal agent of witches’ broom disease of cacao: what’s new from this old foe? Mol Plant Pathol 9:577–588. https://doi.org/10.1111/j.1364-3703.2008.00496.x
Morrissey J, Stack JC, Valls R, Motamayor JC (2019) Low-cost assembly of a cacao crop genome is able to resolve complex heterozygous bubbles. Hortic Res 6:44. https://doi.org/10.1038/s41438-019-0125-7
Motamayor JC, Risterucci AM, Lopez PA, Ortiz CF, Moreno A, Lanaud C (2002) Cacao domestication I: the origin of the cacao cultivated by the Mayas. Heredity (Edinb) 89:380–386. https://doi.org/10.1038/sj.hdy.6800156
Motamayor JC, Lachenaud P, da Silva E, Mota JW, Loor R, Kuhn DN, Brown JS, Schnell RJ (2008) Geographic and genetic population differentiation of the Amazonian chocolate tree (Theobroma cacao L). PLoS ONE 3:e3311. https://doi.org/10.1371/journal.pone.0003311
Motamayor JC, Mockaitis K, Schmutz J, Haiminen N, Livingstone D, Cornejo O, Findley SD, Zheng P, Utro F, Royaert S, Saski C, Jenkins J, Podicheti R, Zhao M, Scheffler BE, Stack JC, Feltus FA, Mustiga GM, Amores F, Phillips W, Marelli JP, May GD, Shapiro H, Ma J, Bustamante CD, Schnell RJ, Main D, Gilbert D, Parida L, Kuhn DN (2013) The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color. Genome Biol 14:r53. https://doi.org/10.1186/gb-2013-14-6-r53
Motilal LA, Zhang D, Mischke S, Meinhardt LW, Boccara M, Fouet O, Lanaud C, Umaharan P (2016) Association mapping of seed and disease resistance traits in Theobroma cacao L. Planta 244:1265–1276. https://doi.org/10.1007/s00425-016-2582-7
Mournet P, Albuquerque PSB de, Alves RM, Silva-Werneck JO, Rivallan R, Marcellino LH, Clément D (2020) A reference high-density genetic map of Theobroma grandiflorum (Willd. ex Spreng) and QTL detection for resistance to witches’ broom disease (Moniliophthora perniciosa). Tree Genet Genomes 16:89. https://doi.org/10.1007/s11295-020-01479-3
Mustiga GM, Gezan SA, Phillips-Mora W, Arciniegas-Leal A, Mata-Quirós A, Motamayor JC (2018) Phenotypic description of Theobroma cacao L. for yield and vigor traits from 34 hybrid families in Costa Rica based on the genetic basis of the parental population. Front Plant Sci 9:808. https://doi.org/10.3389/fpls.2018.00808
Nieves-Orduña HE, Müller M, Krutovsky KV, Gailing O (2021) Geographic patterns of genetic variation among cacao (Theobroma cacao L.) populations based on chloroplast markers. Diversity 13:249. https://doi.org/10.3390/d13060249
Nieves-Orduña HE, Krutovsky KV, Gailing O (2023) Geographic distribution, conservation, and genomic resources of cacao Theobroma cacao L. Crop Sci 63:1750–1778. https://doi.org/10.1002/csc2.20959
Ofori A, Padi FK, Amoako-Attah I (2020) Field evaluation of cacao progenies derived from Guiana clones for yield and black pod disease resistance. Crop Sci 60:249–261. https://doi.org/10.1002/csc2.20101
Orozco-Aguilar L, López-Sampson A, Leandro-Muñoz ME, Robiglio V, Reyes M, Bordeaux M, Sepúlveda N, Somarriba E (2021) Elucidating pathways and discourses linking cocoa cultivation to deforestation, reforestation, and tree cover change in Nicaragua and Peru. Front Sustain Food Syst 5:635779. https://doi.org/10.3389/fsufs.2021.635779
Osorio Montoya T, Henao Ramírez AM, de La Hoz Vasquez T, Urrea Trujillo AI (2022) Propagation of IMC67 plants, universal cacao (Theobroma Cacao L.) rootstock via somatic embryogenesis. Int J Fruit Sci 22:78–94. https://doi.org/10.1080/15538362.2021.2023067
Osorio-Guarín JA, Berdugo-Cely JA, Coronado-Silva RA, Baez E, Jaimes Y, Yockteng R (2020) Genome-wide association study reveals novel candidate genes associated with productivity and disease resistance to Moniliophthora spp. in cacao (Theobroma cacao L.). G3 (Bethesda) 10:1713–1725. https://doi.org/10.1534/g3.120.401153
Paulin D, Ducamp M, Lachenaud P (2008) New sources of resistance to Phytophthora megakarya identified in wild cocoa tree populations of French Guiana. Crop Prot 27:1143–1147. https://doi.org/10.1016/j.cropro.2008.01.004
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Pereira AS, Lopes UV, Pires JL, Barreto MA, Pereira dos Santos L, Gonzaga RS, Da Souza SL, Jesus de Andrade F, Solís Bonilla JL, Gramacho KP (2021) Quantitative resistance to witches’ broom disease in progenies of different sources of resistance. Crop Prot 146:105678. https://doi.org/10.1016/j.cropro.2021.105678
Phillips-Mora W, Castillo J, Krauss U, Rodriguez E, Wilkinson MJ (2005) Evaluation of cacao (Theobroma cacao) clones against seven Colombian isolates of Moniliophthora roreri from four pathogen genetic groups. Plant Pathol 54:483–490. https://doi.org/10.1111/j.1365-3059.2005.01210.x
Phillips-Mora W, Arciniegas-Leal A, Mata-Quirós A, Motamayor JC (2013) Catalogue of cacao clones: selected by CATIE for commercial plantings. Technical series, technical manual, Nr. 105. CATIE, Turrialba, Costa Rica
Ploetz R (2016) The impact of diseases on cacao production: a global overview. In: Bailey BA, Meinhardt LW (eds) Cacao Diseases. Springer International Publishing, Cham, pp 33–59
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
Romero-Navarro JA, Phillips-Mora W, Arciniegas-Leal A, Mata-Quirós A, Haiminen N, Mustiga G, Livingstone D, van Bakel H, Kuhn DN, Parida L, Kasarskis A, Motamayor JC (2017) Application of genome wide association and genomic prediction for improvement of cacao productivity and resistance to black and frosty pod diseases. Front Plant Sci 8:1905. https://doi.org/10.3389/fpls.2017.01905
Royaert S, Jansen J, Silva DV, de Jesus Branco SM, Livingstone DS, Mustiga G, Marelli J-P, Araújo IS, Correa RX, Motamayor JC (2016) Identification of candidate genes involved in Witches’ broom disease resistance in a segregating mapping population of Theobroma cacao L. in Brazil. BMC Genomics 17:107. https://doi.org/10.1186/s12864-016-2415-x
Schnell RJ, Kuhn DN, Brown JS, Olano CT, Phillips-Mora W, Amores FM, Motamayor JC (2007) Development of a marker assisted selection program for cacao. Phytopathol 97:1664–1669. https://doi.org/10.1094/PHYTO-97-12-1664
Schroth G, Läderach P, Martinez-Valle AI, Bunn C, Jassogne L (2016) Vulnerability to climate change of cocoa in West Africa: patterns, opportunities and limits to adaptation. Sci Total Environ 556:231–241. https://doi.org/10.1016/j.scitotenv.2016.03.024
Sereno ML, Albuquerque PSB, Vencovsky R, Figueira A (2006) Genetic diversity and natural population structure of cacao (Theobroma cacao L.) from the Brazilian Amazon evaluated by microsatellite markers. Conserv Genet 7:13–24. https://doi.org/10.1007/s10592-005-7568-0
Singh J, Das S, Jagadis Gupta K, Ranjan A, Foyer CH, Thakur JK (2023) Physiological implications of SWEETs in plants and their potential applications in improving source-sink relationships for enhanced yield. Plant Biotechnol J 21:1528–1541. https://doi.org/10.1111/pbi.13982
Solís Bonilla JL, Vanderlei Lopes U, Zamarripa Colmenero A, Martínez Valencia BB, Avendaño Arrazate CH, Chia Wong JA, Peres Gramacho K (2022) Path analyses define criteria that allow to reduce costs in a breeding population of cacao (Theobroma cacao L.). Tree Genet Genomes 18:25. https://doi.org/10.1007/s11295-022-01554-x
Sosso D, Luo D, Li Q-B, Sasse J, Yang J, Gendrot G, Suzuki M, Koch KE, McCarty DR, Chourey PS, Rogowsky PM, Ross-Ibarra J, Yang B, Frommer WB (2015) Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet 47:1489–1493. https://doi.org/10.1038/ng.3422
Thomas E, van Zonneveld M, Loo J, Hodgkin T, Galluzzi G, van Etten J (2012) Present spatial diversity patterns of Theobroma cacao L. in the neotropics reflect genetic differentiation in Pleistocene refugia followed by human-influenced dispersal. PLoS ONE 7:e47676. https://doi.org/10.1371/journal.pone.0047676
Toxopeus H (1969) Cacao (Theobroma cacao L.). In: Ferwerda F, Wit F (eds) Outlines of perennial crop breeding in the tropics. Veenman, Wageningen
Turnbull C, Hadley P (2023) International Cocoa Germplasm Database (ICGD). [Online Database]. https://www.icgd.reading.ac.uk/index.php. Accessed 24 November 2019
Xie H, Wang D, Qin Y, Ma A, Fu J, Qin Y, Hu G, Zhao J (2019) Genome-wide identification and expression analysis of SWEET gene family in Litchi chinensis reveal the involvement of LcSWEET2a/3b in early seed development. BMC Plant Biol 19:499. https://doi.org/10.1186/s12870-019-2120-4
Zarrillo S, Gaikwad N, Lanaud C, Powis T, Viot C, Lesur I, Fouet O, Argout X, Guichoux E, Salin F, Solorzano RL, Bouchez O, Vignes H, Severts P, Hurtado J, Yepez A, Grivetti L, Blake M, Valdez F (2018) The use and domestication of Theobroma cacao during the mid-Holocene in the upper Amazon. Nat Ecol Evol 2:1879–1888. https://doi.org/10.1038/s41559-018-0697-x
Zhang D, Motilal L (2016) Origin, dispersal, and current global distribution of cacao genetic diversity. In: Bailey BA, Meinhardt LW (eds) Cacao Diseases. Springer International Publishing, Cham, pp 3–31
Zhang D, Figueira A, Motilal L, Lachenaud P, Meinhardt LW (2011) Theobroma. In: Kole C (ed) Wild crop relatives: genomic and breeding resources, vol 6. Springer. Berlin, Heidelberg, pp 277–296
Zhang D, Martínez WJ, Johnson ES, Somarriba E, Phillips-Mora W, Astorga C, Mischke S, Meinhardt LW (2012) Genetic diversity and spatial structure in a new distinct Theobroma cacao L. population in Bolivia. Genet Resour Crop Evol 59:239–252. https://doi.org/10.1007/s10722-011-9680-y
Zhen Q, Fang T, Peng Q, Liao L, Zhao L, Owiti A, Han Y (2018) Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hortic Res 5:14. https://doi.org/10.1038/s41438-018-0024-3
Acknowledgements
The authors would like to thank the International Center for Tropical Agriculture (CATIE) and The International Cocoa Quarantine Centre at the Reading (ICQC, R) for providing the cacao leaf samples for DNA analysis. We acknowledge support by the Open Access Publication Funds of the Georg-August University of Göttingen. We thank anonymous reviewers for insightful suggestions, which helped us improve the manuscript, and editors for handling the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. We thank the German Academic Exchange Service-DAAD for the financial support provided to Helmuth E. Nieves-Orduña. Open access funding was enabled and organized by the German Project DEAL. Funding to OG was provided for the MassArray system by the “Deutsche Forschungsgemeinschaft” (DFG) major research instrumentation grant (reference number: 437098084) and the “Niedersächsisches Ministerium für Wissenschaft und Kultur (MWK) ”.
Author information
Authors and Affiliations
Contributions
HEN-O participated in conceptualization, funding acquisition, investigation, selected samples, performed laboratory work, data analysis, figure elaboration, writing original draft, validation, reviewing, and editing the manuscript; MM participated in data analysis, validation, reviewing, and editing the manuscript; KVK participated in conceptualization, investigation, data analysis, supervision, validation, reviewing, and editing the manuscript; OG participated in conceptualization, funding acquisition, investigation, data analysis, project administration, supervision, validation, reviewing, and editing the manuscript. All authors commented on and edited intermediate versions of the manuscript. All authors reviewed and approved the final manuscript.
Corresponding authors
Additional information
Communicated by C. Chen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nieves-Orduña, H.E., Müller, M., Krutovsky, K.V. et al. Genotyping of cacao (Theobroma cacao L.) germplasm resources with SNP markers linked to agronomic traits reveals signs of selection. Tree Genetics & Genomes 20, 13 (2024). https://doi.org/10.1007/s11295-024-01646-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-024-01646-w