Abstract
Cork oak (Quercus suber L.) is an ecologically and economically important evergreen tree species native to the Mediterranean region and widespread in southwest Europe and northwest Africa. An improved genome assembly of cork oak using a combination of Illumina and PacBio sequencing is presented in this study. The assembled genome contains 2351 scaffolds longer than 1000 bp, accounting for 765.7 Mbp of genome size, L90 of 755, and a N50 of 1.0 Mbp, with 40,131 annotated genes. The repetitive sequences constitute 53.6% of the genome. The genome sequences of chloroplast and mitochondrion were determined for the first time, with a genome size of 161,179 bp and 531,858 bp, respectively. Phylogenetic analysis based on complete chloroplast genome sequence showed that Q. suber is closely related to Quercus variabilis, two cork-producing species with commercial use. All data generated are available through the public databases, being ready to be used without restrictions. This study provides an improved nuclear genome assembly together with the organelle genomes of cork oak. These resources will be useful for further breeding strategies and conservation programs and for comparative genomic studies in oak species.
Similar content being viewed by others
Introduction
Cork oak (Quercus suber L.) is a forest tree species native to the Mediterranean region with a significant economic importance. Cork trees are found in human managed cork oak woodlands (known as “montado” in Portugal, and “dehesa” in Spain); these are important habitats harboring a wide range of biodiversity, including many rare, endemic, and endangered plant and animal species (Berrahmouni et al. 2009). The long lifespan and high activity of the cork cambium in this species allow the sustainable exploitation of its outer bark (cork) for a wide range of industrial applications (Leite and Pereira 2017), harnessing its unique physical and chemical properties.
The Mediterranean region is considered one of the most affected by climate change, and the intensification of extreme climatic events is imposing a significant pressure on cork productivity and tree survival (Nunes et al. 2021; Pérez-Girón et al. 2022). Events of sudden death or progressive decline have long been reported in cork oak and are thought to become more frequent due to the combined occurrence of extreme drought coupled with biotic stress (Camilo-Alves et al. 2017). Recent advances in silviculture practices have been proposed as a way to minimize the impact of these extreme events on tree development in short term (Camilo-Alves et al. 2020); however, a comprehensive knowledge of the cork oak genetic heritage is critical to plan long-term strategies for plant improvement.
The draft nuclear genome of cork oak was made available in 2018 (Ramos et al. 2018), providing a reference resource for genomic studies in this species, and the third genome assembly available for the Quercus genus (Sork et al. 2016; Plomion et al. 2018). The three genome assemblies revealed a high similarity, despite the long evolutionary distance (approx. 35 MYA) from a common ancestor (Hipp et al. 2020; Sork et al. 2022). The draft cork oak genome assembly was produced using Illumina paired-end (PE) and mate-pair (MP) sequencing. It was fragmented in 23,344 scaffolds, with 94.6% of the genome represented in 4730 scaffolds larger than 10 Kbp, including 79,752 genes with complete open reading frames (Ramos et al. 2018). The predicted size was 953.3 Mbp in close agreement with previous estimates using flow cytometry (Zoldos et al. 1998).
The availability of a reference genome for cork oak paved the way for multiple transcriptomic studies aimed at unveiling key molecular mechanisms regulating secondary growth (Leal et al. 2021) and cork development (Lopes et al. 2020; Fernández-Piñán et al. 2021; Pires et al. 2022). Using additional transcriptomic datasets already available for different tissues, which included EST sequencing (Pereira-Leal et al. 2014), other authors focused on genome-wide or targeted characterization of genes involved in epigenetic regulation (Inácio et al. 2018; Silva et al. 2020), triterpenoid synthesis (Busta et al. 2020), and response to biotic stress (Coelho et al. 2021). In addition, the development of genetic markers for early selection of more productive and resilient genotypes would be extremely useful for this species, and different authors have started to explore the occurrence of genetic diversity associated with cork quality (Mendes et al. 2022) and adaptation to climate change (Vanhove et al. 2021).
In the present paper, we describe the development of the most recent version of the cork oak genome, obtained using a combined assembly of Illumina and PacBio sequencing reads. We also report for the first time the sequence assembly of the cork oak plastidial and mitochondrial genomes.
Materials and methods
Plant material and DNA extraction
Cork oak leaves were collected from the same HL8 tree used to generate the draft version of the cork oak reference genome assembly (Ramos et al. 2018) in order to produce long-read sequences. This specimen is over 50 years old and considered a producer of very high-quality cork. Total nuclear DNA extraction of the newly collected leaves was obtained with the NGS-Grade gDNA Prep optimized for PacBio long reads and performed by Amplicon Express (USA). Additional leaves were also submitted to organelle DNA isolation using the protocol described by Lang et al. (2011). The innuPREP Plant DNA Kit (Analytik Jena, Jena, DE) was used for DNA extraction.
Sequencing
The nuclear DNA sequencing was carried out using the PacBio RS II platform at the Yale Center for Genomic Analysis, Yale University (USA). Organelle DNA sequencing was performed using the Illumina HiSeq 4000 platform, at the Beijing Genomics Institute (BGI) producing PE reads of 100 bp in length with an insert size of 300 bp.
Moreover, the Illumina PE and MP reads and the RNA-Seq data produced to build the draft genome version of cork oak were also used in this work (Supplementary Table 1).
Illumina raw reads obtained from the nuclear DNA libraries and the RNA-Seq reads were preprocessed with Trimmomatic v.0.36 (Bolger et al. 2014). Adapter sequences and low-quality bases with less than an average quality threshold of 20 over a sliding window of 10% of the read length were trimmed from the end of the read; reads shorter than 80% of the original read length were then removed. Finally, the PacBio long raw reads were self-corrected using Canu v1.5 with the following parameter specification: rawErrorRate=0.25, correctedErrorRate=0.04, corOutCoverage=90, and userGrid=false (Koren et al. 2017).
The RNA-Seq reads were preprocessed with Trimmomatic using the same criteria described above.
Genome assembly and annotation
To remove chloroplast and mitochondrial sequencing reads, the preprocessed reads were mapped against the Quercus suber chloroplast and mitochondrial genomes assembled during this work and described further below, using the BWA-mem algorithm (Li 2013). Unmapped reads from such alignments were used for downstream assembly of the nuclear genome.
The bioinformatics pipeline followed is described in Fig. 1. As a first step, two independent genome assemblies based on short reads were produced using Ray assembler (Boisvert et al. 2010). While a k-mer size of 81 bp was defined for the assembly based on reads of 100 bp, a k-mer size of 121 bp was set for the assembly based on reads of 150 bp. Possible alternative heterozygous sequences were removed from each assembly using Redundans (Pryszcz and Gabaldón 2016). Then, assembly reconciliation was performed with GARM (Soto-Jimenez et al. 2014) as previously described (Ramos et al. 2018).
The MP reads were mapped against the final set of contigs obtained from the assembly reconciliation with BWA-mem. Only the reads with a minimum mapping quality score of 10 were subsequently used for the scaffolding process using the SSPACE Basic (Boetzer et al. 2011), with the MP libraries used in ascending order relative to their respective insert size. In order to reduce the number of gaps of the genome, a first round of gap closing was performed using the long error-corrected reads of PacBio with the SOAPdenovo2 (Luo et al. 2012). Afterwards, three additional rounds of (1) gap closing with two PE libraries from all available insert sizes (170 bp, 300 bp, 500 bp, and 800 bp), (2) scaffolding using the long error-corrected PacBio reads with the SSPACE-LongRead (Boetzer and Pirovano 2014), and (3) scaffolding with the MP using SSPACE Basic were performed. In each round, the set of PE libraries used for the gap closing step was different (Fig. 1, red square). Lastly, a screening for potential contaminants was performed with FCS-GX from the Foreign Contamination Screening (FCS) tool (https://github.com/ncbi/fcs).
The gene prediction of the cork oak genome was performed executing two rounds of annotation with Maker (Campbell et al. 2014), using several sources of external hints generated to maximize annotation accuracy (see Supplementary Materials). In the first round, Maker was run to predict gene coordinates based on the evidences provided from all the hints generated. The set of genes obtained from the first round was used to train Augustus and SNAP (Korf 2004; Stanke et al. 2008). Once both kinds of software were trained, the second round of Maker was performed in order to do an ab initio gene prediction keeping the predictions made in the first round.
Functional annotation of the final set of genes was performed using BLAST (Camacho et al. 2009), against the NCBI non-redundant plants and the Swiss-Prot databases (Boeckmann et al. 2003). Additionally, conserved protein domains, Gene Ontology (GO) terms, and KEGG mappings were identified using InterProScan (Jones et al. 2014). Also, eggNOG-mapper (Huerta-Cepas et al. 2017) was used to assign orthologues based on precomputed Viridiplantae phylogenies.
The quality of the annotation was assessed by the annotation edit distance (AED) metric which quantifies the congruency between a gene annotation and its supporting evidence.
Transposable and repetitive elements (TEs and REs)
The first set of TEs and REs was identified and annotated running RepeatMasker v.4.0 (Smith et al. 2013) using the eudicotyledons subset of RepBase. Additionally, due to the highly variable sequences of TEs across species and to accurately identify the diverse set of TEs distributed in the cork oak genome sequence, RepeatModeler2 (Flynn et al. 2020) was used to produce a de novo reference library of TEs.
Completeness of the genome assembly
CEGMA and BUSCO v5.3.0 (Parra et al. 2007; Manni et al. 2021) were used to investigate the presence of highly conserved orthologous genes in the nuclear genome assembly. CEGMA contains a total of 248 core eukaryotic genes. BUSCO was run over the plant set (embryophyta_odb10), which includes a total of 1614 ortholog groups, defining Arabidopsis thaliana as the model species for the gene prediction performed by Augustus within the BUSCO pipeline.
Chloroplast genome assembly and annotation
For the chloroplast (cp) genome assembly, NOVOPlasty v2.6.3 (Dierckxsens et al. 2017) was used, setting insert Range and Insert Range strict parameters to 1.3 and 1.1, respectively. The complete CDS sequence of Quercus suber ribulose 1,5-bisphosphate carboxylase/oxygenase large subunit (GenBank: AB125027.1) was used as a seed for the de novo assembly of the cp genome, following software recommendations. By using a conserved organelle gene sequence as a seed to start the assembly, followed by sequence extension, the evidence of genome circularization is better achieved than in more common genome assembly strategies, which prioritize the assembly of the entire dataset at once. Furthermore, given that organelles have homologous sequences, a more targeted approach like the one presented here may reduce the cross contamination of sequences between organelles during genome assembly. Additionally, the complete cp genome of Quercus variabilis (GenBank: NC_031356.1, (Yang et al. 2016)) was used as a reference genome to resolve highly repetitive regions.
The cp genome sequence was then blasted against NCBI non-redundant database to confirm its integrity and completeness compared with other Quercus cp genomes available.
The cp genome was annotated using the web-based tool DOGMA (Wyman et al. 2004) with default parameters. Results were manually inspected, and the precise coordinates of each gene were confirmed using the annotation of Q. variabilis as a guide. For genes not automatically detected, but present in Q. variabilis annotation, a manual search for Q. variabilis gene sequence was performed, and the presence of the gene was then confirmed or discarded. New putative genes present in our annotation, but not in Q. variabilis, were verified by homology with chloroplast proteins from other species through blastx in NCBI non-redundant database.
Thirty-nine complete cp genome sequences were used for phylogenetic analysis, including the new chloroplast genome of Q. suber and 35 cp genomes of different Quercus species publicly available. Arabidopsis thaliana (NC_000932), Populus trichocarpa (NC_009143), and Fagus sylvatica (NC_041437) were used as outgroup species. The details of the Quercus species used are provided in Supplementary Table 2. The 39 cp genomes were aligned with MAFFT v.7 with default parameters (Katoh et al. 2018). The alignment was trimmed using trimAl v1.2rev22 (Capella-Gutiérrez et al. 2009) with options -cons 60 and -gt 0.9 and manually adjusted. Phylogenetic analysis was conducted by maximum likelihood (ML) using RAxML v8.2.12 (Stamatakis 2014) with the GTRGAMMA model and 1000 bootstrap replicates. The tree was rooted at midpoint.
Mitochondrial genome assembly and annotation
The mitochondrion (mt) genome was assembled following the same approach described for the chloroplast genome, using NOVOPlasty. However, in this case, the partial CDS sequence of Orfx gene (GenBank: DQ340806.1) was used to initiate the assembly. The complete mt genome of Betula pendula (GenBank: LT855379.1) was used as reference to resolve highly repetitive regions. Furthermore, the chloroplast genome obtained previously was also considered in order to exclude chloroplast reads that could be present in the data. To reduce the fragmentation of the genome, a de novo assembly based only on the organelle-unfiltered error-corrected PacBio reads was performed with Canu v.1.5 (for more details see Supplementary Materials). Then, the contigs obtained from NOVOPlasy were aligned with the PacBio assembly using LAST v885 (Kiełbasa et al. 2011). Additionally, the preprocessed MP reads were mapped with BWA-mem against the improved mt genome assembly to search for evidences of junction between contigs. Both organelle genomes were also aligned against each other with LAST to check for cross contamination of sequences.
MITOFY analysis web server (Alverson et al. 2010) was used with default parameters to find protein coding and rRNA genes for Q. suber mitochondrial genome. This annotation was then manually confirmed gene by gene in the web server by comparison with other land plant species available at MITOFY database. Transfer RNA (tRNA) genes were detected using tRNAscan-SE tool (Lowe and Chan 2016) with default parameters.
Results and discussion
Nuclear genome assembly and annotation
The percentage of reads kept after preprocessing ranged from 56.2 to 93.3% (Table 1). The Illumina preprocessed reads were assembled separately by their original read length, 100 and 150 bp, resulting in two assemblies with 249,707 and 214,524 contigs, respectively. Then, a haplotype reduction was performed over each assembly which allowed for a reduction of the number of contigs by 55.50% and 56.74%, respectively. The assembly reconciliation procedures were applied at the end producing a single assembly comprising 111,877 contigs. More details about the assemblies are provided in Supplementary Table 3. These contigs were then used as the basis of the scaffolding and gap filling processes with MP and corrected PacBio reads, producing a final genome assembly with 2479 scaffolds. The assembly statistics through the scaffolding and gap filling procedures are shown in Table 2. Lastly, after screening for potential contaminants, 128 sequences were removed due to a high homology with bacterial and fungal genomes.
The final assembly of cork oak genome contained 2351 scaffolds greater than 1000 bp, representing an assembly length of 765.7 Mbp with a 4.3% of undetermined nucleotides. The N50 obtained was 1.0 Mbp, and the largest scaffold was 4,440,151 bp in length. Most of the genome was represented in the largest scaffolds. For instance, scaffolds equal to or larger than 250 Kbp (N=823) comprise approximately 92.5% of the genome (Table 3), being the L90 equal to 755.
Using two rounds of an automated annotation pipeline based on Maker, 40,131 genes were predicted (23,512 in the first round). The full set of predicted genes represented 21.1% of the total genome size, with a density of 0.52 genes/10 kb, which is similar to the densities of Quercus robur (GCA_900291515) and Quercus lobata (GCA_001633185) (0.32 and 0.63 genes/10 kb, respectively), two species of the same genus and similar genome size (814.3 Mbp and 845.94 Mbp, respectively) (Plomion et al. 2018; Sork et al. 2022).
From the whole set of predicted genes, 33,006 (82.2%) and 39,504 (98.4%) were functionally annotated against Swiss-Prot and NCBI-nr plant databases, respectively. Additionally, the search for protein signatures against InterPro resulted in 37,184 (92.6%) genes with similarity to at least one protein domain. Regarding the GO terms and KEGG pathways, 16,198 (40.3%) genes were assigned to at least one GO term and 2727 (6.8%) genes to an Enzyme Commission Number.
The annotation edit distance (AED) was used to evaluate the annotation quality of the cork oak genome. AED values range from 0 to 1, with 0 denoting an extrinsic evidence and 1 denoting no extrinsic evidence supporting the annotation (Campbell et al. 2014). Figure 2 shows the cumulative distribution of AED values of the predicted genes. A total of 38,513 genes (95.9%) have an AED < 0.5, and 27,409 (68.3%) have at least one recognizable Pfam protein domain. Based on the rule of thumb defined by Campbell et al. (Campbell et al. 2014), the cork oak genome annotation can be considered well annotated.
CEGMA and BUSCO results
The results obtained by CEGMA showed that the completeness of the genome was 98.8% (98.4% of complete matches) while with BUSCO, it was 91.6% (88.6% of complete matches). Comparing these results against the results obtained in the draft version of the cork oak genome (Ramos et al. 2018), the completeness reported by CEGMA remains the same, while the one reported by BUSCO was slightly lower in the current version. Although the standard metrics suggested that duplicate orthologues were better resolved in this version (6.8% vs. 10.4%), the number of missing orthologues increased. This increase could be a fallout of the application of heterozygous sequence reduction, process from which Pryszcz and Gabaldón (2016) emphasize that the resulting assembly represents a variety of fragments which are randomly chosen from each of the haploid genomes.
Genome size and gene representation
To better understand the difference in the cumulative size of the genome between the draft version and the genome version reported here, which is about 187.6 Mb smaller in size, both genomes were aligned with LAST using the current version as reference. The alignment results showed that 95.4% of the current genome sequence is represented in the draft version, with 2337 scaffolds out of 2351 showing alignments with 22,877 scaffolds from the draft genome. A total of 17.5 Kbp of sequence was recovered from the current genome version and not assembled in the draft version. The incorporation of the long PacBio reads allowed the capture of more challenging sequence architecture. Additionally, together with the removal of possible alternative heterozygous sequences immediately after the assembly allowed improving the scaffolding rearrangement. However, 65.8 Kbp of the draft sequence is not present in the current version of the cork oak genome. This could be directly related to the smaller genome size and slightly higher number of fragmented and missing ortholog genes, as a result of different methodologies applied for genome assembly.
Regarding the number of genes predicted (and gene content), the gene sequences from the draft were blasted against the current genome at the nucleotide level, with an e-value of 1e−5. From the 79,372 genes, 70.0% (55,627) aligned against the current genome version, although only 45.7% aligned in regions with annotated genes. It should be kept in mind that fragmented assemblies lead to an overestimated number of annotated genes (Denton et al. 2014). In fact, the number of high-confidence genes identified in the draft version (33,658) is closer to the number of genes predicted in the updated version (40,131).
Transposable and repetitive elements
The percentage of TEs and REs covering the genome based on RepBase was 11.6%, for a total of 88,930,547 bp of the 765.7 Mbp (732.8 Mbp excluding Ns). Retroelements account for 6.9% of the genome while DNA transposons and simple repeats covered 0.8% and 2.9%, respectively (Supplementary Table 4). Similar results were found in the draft version of the genome (Ramos et al. 2018). Additionally, to better assess the tandem repeats present in the cork oak genome, the modeling step based on the combination of RepeatModeler2 + RepeatMasker was essential. With this strategy, we could predict a level of repetitiveness of 53.4%, in agreement with the level reported for Quercus mongolica (53.7%) (Ai et al. 2022) and other Quercus species (Sork et al. 2022) (Supplementary Table 5).
Chloroplast genome
The chloroplasts are the plant organelles responsible for the conversion of light energy into chemical energy through the process of photosynthesis (Finkeldey and Gailing 2013). The cp genome presents inheritance patterns, predominantly of maternal nature (Greiner et al. 2015), and its characterization has been useful for evolutionary studies and phylogenetic relationships (Wu et al. 2020). In this study, we assembled and characterized the complete cp genome sequence of Q. suber.
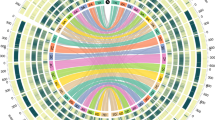
From the initial 13,519,700 whole-genome Illumina PE reads, only 274,918 (2.03%) were de novo assembled as cp genome, representing an average coverage depth of 253x. This assembly yielded one single circular molecule of 161,179 bp (Fig. 3), which is among the range of sizes known for the thirty five cp genomes of Quercus species (from 160,415 to 161,394 bp, in Quercus aquifolioides and Quercus bawanglingensis, respectively), available at NCBI organelle genome Resources (https://www.ncbi.nlm.nih.gov/genome/organelle/) and summarized in Supplementary Table 2. The GC content was 36.8%, also very similar to other Quercus, which range between 36.79 and 36.96% depending on the species. The cork oak cp genome annotation presented 90 protein coding genes, 40 tRNA and 8 rRNA genes. The comparison of these results with the annotation of five cp genomes reported by Yang et al. (2016) reveals that the annotation of Q. suber cp genome is in complete agreement with other Quercus species.
Circular map of cork oak chloroplast genome. The color of the genes corresponds to their function as per the legend. Genes transcribed counter-clockwise are indicated inside of the outer circle, while genes transcribed clockwise are displayed outside of the circle. The inner circle represents the GC content as a histogram in darker gray. The gray line in the inner circle represents 50% GC content. The black border line of the inner circle represents the different regions of chloroplast genome (IRA, IRB: inverted repeats; SSC: small single copy; LSC: large single copy). This plot was generated with OGDRAW tool (Greiner et al. 2019)
Quercus species have been classified into two main lineages known as the “New World” and the “Old World” oak clades (Manos et al. 1999). The “New World” oak clade is formed by the subgenus Quercus which comprises five sections (Quercus, Lobatae, Protobalanus, Ponticae, and Virentes), while the “Old World” oak clade is formed by the subgenus Cerris comprised by three sections (Cerris, Cyclobalanospsis, and Ilex). Considering this classification, the phylogenetic tree based on the complete cp genomes showed that the Quercus section formed a single clade. The Ilex section was split into two clusters. The first cluster is formed by species distributed across Tibet, Nepal, Bhutan, Myanmar, and Thailand regions (Q. spinose, Q. tungmaiensis, Q. aquifolioides, and Q. pannosa) together with Q. guyavifolia, present in nearby China. The second one is comprised by Quercus species occurring in Japan and Southern and Central China. The exceptions were Q. baronii and Q. acrodonta that appeared to be closely related to section Cerris, where Q. suber belongs. This reflects the monophyletic relation between Cerris and Ilex, which was well established at the nuclear level before (Denk and Grimm 2010). Cork oak grouped relatively close to Q. variabilis, both cork-producing species for commercial use. Additionally, the sections from the “Old Word” are clearly distinct from the clade comprising Quercus, Lobatae, and Virinetes sections (Fig. 4, Supplementary Table 2).
Phylogenetic tree obtained for 39 complete chloroplast genomes, 36 corresponding to Quercus species. Black circle: section Cerris; black square: section Ilex; black triangle: section Cyclobalanospsis; black rhombus: section Quercus; white circle: section Lobatae; white square: section Virentes; white rhombus: outgroup species; nothing: undetermined section
Mitochondrial genome
A total of 612,018 out of 11,269,248 whole-genome Illumina PE reads (5.43%) were assembled into 23 contigs, from which 16 showed evidence of overlap. The contig sizes varied from 1123 to 81,375 bp, for a total genome length of 796,392 bp. The self-alignment of these 23 contigs showed high homology between different contigs, which suggested (1) duplicate regions of mt genome or (2) a poor assembly by the NOVOPlasty software. In order to improve these results, we took advantage of a de novo assembly of Q. suber nuclear genome previously described in this work, which used error-corrected PacBio reads unfiltered for organelle contamination. The alignment of this PacBio assembly against the contigs obtained with NOVOPlasty identified 12 PacBio contigs with high homology. The manual inspection of the alignment coordinates allowed the overlap of several PacBio contigs after confirming sequence identity. Three contigs were obtained for mitochondrion assembly: one large contig with 442,094 bp, followed by two smaller contigs of 52,064 bp and 37,700 bp. The MP reads of different insert sizes were further mapped to this assembly, but no evidence of connections between contigs was found. These contigs, which showed no evidence of circularization, sum a total genome length of 531,858 bp. The genome length is larger than other Quercus mitochondrion genomes recently deposited, namely, Q. robur (390,906 bp GenBank: OW028777.1), Q. variabilis (412,886 bp; Genbank: MN199236.1), and Q. acutissima (448,694; GenBank: MZ636519.1). The annotation of Q. suber mt genome revealed a total of 66 genes, being 40 protein coding, 23 tRNA, and 3 rRNA. The completeness of the annotation is corroborated by the presence of all major organelle genes except for RPS9 and RPS13. This annotation presents more genes than any other Quercus mitochondrion genome, which is in agreement with its increased genome length.
Comparison with other Quercus genomes
Our assembly was compared against five other available genomes for Quercus genus (Table 4, data accessed on 30/05/2022). The L90 metric, which describes the minimum number of sequences representing 90% of the genome sequence, is expected to be equal or closer to the number of chromosomes when genome assembly contiguity is very high, being a good method to better compare and assess genome contiguity. Four genomes had an L90 between 11 and 12 and were organized in chromosomes (Q. aquifolioides, Q. lobata, Q. robur, Q. mongolica, and Q. variabilis) and one in scaffolds (Quercus wislizeni). All assemblies were aligned against the cork oak genome using LAST v885. The alignment results revealed a high similarity between Q. suber and the other Quercus species, with 98.16% of Q. aquifoloides, 96.70% of Quercus wisilizeni, 95.94% of Q. robur, 95.72% of Q. mongolica, and 95.43% of Q. lobata genomes, aligned against the cork oak genome.
BUSCO analysis
The Q. suber genome reported here is slightly less complete than the other genomes represented in Table 5 (Q. robur v.3.1 (98.0%); Q. wislizeni (97.7%); Q. lobata (96.8%); Q. mongolica (95.4%), Q. aquifolioides (95.1%); and Q. suber (88.6%)) but still has a high-level of completeness.
Conclusions
High-quality genome assemblies are of great need, since initial “drafts” may fail to capture important genomic regions involved in specific adaptations of the species. In this work, the addition of long-sequences from PacBio allowed to improve sequence connectivity and reduce fragmentation of the cork oak reference genome (Ramos et al. 2018), observing a high level of sequence assembly homology between the two versions. However, genome completeness was slightly lower mostly due to the differences in the pipeline applied which resulted in a smaller genome size.
Another important goal of this study was the assembly of the cork oak plastidial and mitochondrial genomes, which was accomplished for the first time. These newly reported genomes will provide valuable information for genetic evolution and molecular breeding studies of Quercus species.
Given the importance of cork oak and the efforts required to study its genetic structure, it is essential to be able to use all the information resources available. Therefore, the current version of the cork oak genome will be deposited in the CorkOakDB (Arias-Baldrich et al. 2020) to integrate and share open access to the information in a comprehensive, accessible, and intuitive format.
High-quality genomes allow the documenting of the deep evolutionary history of species. The current cork oak genome version is a step forward towards a better and less fragmented genome sequence, and further improvements can be achieved making use of recent sequence technologies such as HiFi (high fidelity) or Hi-C (high-throughput chromosome conformation capture technique) sequencing data. Hence, our research team has additional ongoing work to develop a better version of the cork oak genome at the chromosome level, which will lead to additional improvements in the quality and completeness of the cork oak genome.
Data availability
The raw sequencing data and the nuclear and organelle genomes presented in this study are deposited in the NCBI Sequence Read Archive under the BioProject ID PRJNA392919.
References
Ai W, Liu Y, Mei M et al (2022) A chromosome-scale genome assembly of the Mongolian oak (Quercus mongolica). Mol Ecol Resour 22:2396–2410. https://doi.org/10.1111/1755-0998.13616
Alverson AJ, Wei X, Rice DW et al (2010) Insights into the evolution of mitochondrial genome size from complete sequences of citrullus lanatus and cucurbita pepo (Cucurbitaceae). Mol Biol Evol 27:1436–1448. https://doi.org/10.1093/molbev/msq029
Arias-Baldrich C, Silva MC, Bergeretti F et al (2020) CorkOakDB—the Cork Oak Genome Database Portal. Database 2020. https://doi.org/10.1093/database/baaa114
Berrahmouni N, Regato P, Ellatifi M et al (2009) Ecoregional planning for biodiversity conservation. Cork oak woodlands edge Isl Press, Washington, USA, pp 203–216
Bodénès C, Chancerel E, Ehrenmann F et al (2016) High-density linkage mapping and distribution of segregation distortion regions in the oak genome. DNA Res An Int J Rapid Publ Rep Genes Genom 23:115. https://doi.org/10.1093/DNARES/DSW001
Boeckmann B, Bairoch A, Apweiler R et al (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31:365–370. https://doi.org/10.1093/NAR/GKG095
Boetzer M, Henkel CV, Jansen HJ et al (2011) Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. https://doi.org/10.1093/bioinformatics/btq683
Boetzer M, Pirovano W (2014) SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinform 15:211. https://doi.org/10.1186/1471-2105-15-211
Boisvert S, Laviolette F, Corbeil J (2010) Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 17:1401–1415. https://doi.org/10.1089/cmb.2009.0238
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/BIOINFORMATICS/BTU170
Busta L, Serra O, Kim OT et al (2020) Oxidosqualene cyclases involved in the biosynthesis of triterpenoids in Quercus suber cork. Sci Rep 10:1–12. https://doi.org/10.1038/s41598-020-64913-5
Camacho C, Coulouris G, Avagyan V et al (2009) BLAST+: architecture and applications. BMC Bioinform 10:1–9. https://doi.org/10.1186/1471-2105-10-421
Camilo-Alves C, Dinis C, Vaz M et al (2020) Irrigation of young cork oaks under field conditions—testing the best water volume. Forests 11:88. https://doi.org/10.3390/f11010088
Camilo-Alves CSP, Vaz M, Da Clara MIE, Ribeiro NMDA (2017) Chronic cork oak decline and water status: new insights. New For 48:753–772. https://doi.org/10.1007/s11056-017-9595-3
Campbell MS, Holt C, Moore B, Yandell M (2014) Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinform 2014:4.11.1–4.11.39. https://doi.org/10.1002/0471250953.bi0411s48
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. https://doi.org/10.1093/BIOINFORMATICS/BTP348
Coelho AC, Pires R, Schütz G et al (2021) Disclosing proteins in the leaves of cork oak plants associated with the immune response to Phytophthora cinnamomi inoculation in the roots: a long-term proteomics approach. PLoS One 16:e0245148. https://doi.org/10.1371/journal.pone.0245148
Denk T, Grimm GW (2010) The oaks of western Eurasia: traditional classifications and evidence from two nuclear markers. Taxon 59:351–366. https://doi.org/10.1002/TAX.592002
Denton JF, Lugo-Martinez J, Tucker AE et al (2014) Extensive error in the number of genes inferred from draft genome assemblies. PLoS Comput Biol 10:1003998. https://doi.org/10.1371/JOURNAL.PCBI.1003998
Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res 45:e18. https://doi.org/10.1093/nar/gkw955
Fernández-Piñán S, Boher P, Soler M et al (2021) Transcriptomic analysis of cork during seasonal growth highlights regulatory and developmental processes from phellogen to phellem formation. Sci Rep 11:1–14. https://doi.org/10.1038/s41598-021-90938-5
Finkeldey R, Gailing O (2013) Chloroplasts. In: Brenner’s encyclopedia of genetics, 2nd edn. Elsevier Inc., pp 525–527
Flynn JM, Hubley R, Goubert C et al (2020) RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A 117:9451–9457. https://doi.org/10.1073/PNAS.1921046117/
Greiner S, Lehwark P, Bock R (2019) OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res 47:W59. https://doi.org/10.1093/NAR/GKZ238
Greiner S, Sobanski J, Bock R (2015) Why are most organelle genomes transmitted maternally? BioEssays 37:80–94. https://doi.org/10.1002/bies.201400110
Hipp AL, Manos PS, Hahn M et al (2020) Genomic landscape of the global oak phylogeny. New Phytol 226:1198–1212. https://doi.org/10.1111/nph.16162
Huerta-Cepas J, Forslund K, Coelho LP et al (2017) Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34:2115–2122. https://doi.org/10.1093/molbev/msx148
Inácio V, Martins MT, Graça J, Morais-Cecílio L (2018) Cork oak young and traumatic periderms show pcd typical chromatin patterns but different chromatin-modifying genes expression. Front Plant Sci 9:1194. https://doi.org/10.3389/fpls.2018.01194
Jones P, Binns D, Chang HY et al (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. https://doi.org/10.1093/bioinformatics/btu031
Katoh K, Rozewicki J, Yamada KD (2018) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. https://doi.org/10.1093/bib/bbx108
Kiełbasa SM, Wan R, Sato K et al (2011) Adaptive seeds tame genomic sequence comparison. Genome Res 21:487–493. https://doi.org/10.1101/gr.113985.110
Koren S, Walenz BP, Berlin K et al (2017) Canu: Scalable and accurate long-read assembly via adaptive κ-mer weighting and repeat separation. Genome Res 27:722–736. https://doi.org/10.1101/gr.215087.116
Korf I (2004) Gene finding in novel genomes. BMC Bioinform 5:59. https://doi.org/10.1186/1471-2105-5-59
Lang EGE, Mueller SJ, Hoernstein SNW et al (2011) Simultaneous isolation of pure and intact chloroplasts and mitochondria from moss as the basis for sub-cellular proteomics. Plant Cell Rep 30:205–215. https://doi.org/10.1007/s00299-010-0935-4
Leal AR, Sapeta H, Beeckman T et al (2021) Spatiotemporal development of suberized barriers in cork oak taproots. Tree Physiol. https://doi.org/10.1093/treephys/tpab176
Leite C, Pereira H (2017) Cork-containing barks—a review. Front Mater 3:63. https://doi.org/10.3389/fmats.2016.00063
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997. https://doi.org/10.48550/arXiv.1303.3997
Lopes ST, Sobral D, Costa B et al (2020) Phellem versus xylem: genome-wide transcriptomic analysis reveals novel regulators of cork formation in cork oak. Tree Physiol 40:129–141. https://doi.org/10.1093/treephys/tpz118
Lowe TM, Chan PP (2016) tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44:W54–W57. https://doi.org/10.1093/nar/gkw413
Luo R, Liu B, Xie Y et al (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. https://doi.org/10.1186/2047-217X-1-18
Manni M, Berkeley MR, Seppey M, Zdobnov EM (2021) BUSCO: assessing genomic data quality and beyond. Curr Protoc 1:e323. https://doi.org/10.1002/cpz1.323
Manos PS, Doyle JJ, Nixon KC (1999) Phylogeny, biogeography, and processes of molecular differentiation in Quercus subgenus Quercus (Fagaceae). Mol Phylogenet Evol 12:333–349. https://doi.org/10.1006/MPEV.1999.0614
Mendes B, Usié A, Capote T et al (2022) Quercus suber transcriptome analyses: identification of genes and SNPs related to cork quality. In: Biology and Life Sciences Forum 2022, vol 11. MDPI AG, p 76. https://doi.org/10.3390/IECPS2021-11916
Nunes LJR, Meireles CIR, Gomes CJP, Ribeiro NMCA (2021) The impact of climate change on forest development: a sustainable approach to management models applied to Mediterranean-type climate regions. Plants 11:69. https://doi.org/10.3390/plants11010069
Parra G, Bradnam K, Korf I (2007) CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. https://doi.org/10.1093/bioinformatics/btm071
Pereira-Leal JB, Abreu IA, Alabaça CS et al (2014) A comprehensive assessment of the transcriptome of Cork oak (Quercus suber) through EST sequencing. BMC Genom 15:1–14. https://doi.org/10.1186/1471-2164-15-371
Pérez-Girón JC, Díaz-Varela ER, Álvarez-Álvarez P (2022) Climate-driven variations in productivity reveal adaptive strategies in Iberian cork oak agroforestry systems. For Ecosyst 9:100008. https://doi.org/10.1016/j.fecs.2022.100008
Pires RC, Ferro A, Capote T et al (2022) Laser microdissection of woody and suberized plant tissues for RNA-Seq analysis. Mol Biotechnol 1–14. https://doi.org/10.1007/S12033-022-00542-9
Plomion C, Aury JM, Amselem J et al (2018) Oak genome reveals facets of long lifespan. Nat Plants 4:440–452. https://doi.org/10.1038/s41477-018-0172-3
Pryszcz LP, Gabaldón T (2016) Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res 44:e113. https://doi.org/10.1093/nar/gkw294
Ramos AM, Usié A, Barbosa P et al (2018) The draft genome sequence of cork oak. Sci Data 5:1–12. https://doi.org/10.1038/sdata.2018.69
Silva HG, Sobral RS, Magalhães AP et al (2020) Genome-wide identification of epigenetic regulators in Quercus suber L. Int J Mol Sci 21:3783. https://doi.org/10.3390/ijms21113783
Smith A, Hubley R, Green P (2013) RepeatMasker Open-4.0. RepeatMasker Open-40
Sork VL, Cokus SJ, Fitz-Gibbon ST et al (2022) High-quality genome and methylomes illustrate features underlying evolutionary success of oaks. Nat Commun 13:1–15. https://doi.org/10.1038/s41467-022-29584-y
Sork VL, Fitz-Gibbon ST, Puiu D et al (2016) First draft assembly and annotation of the genome of a California endemic oak Quercus lobata Née (Fagaceae). G3 Genes|Genom|Genet 6:3485–3495. https://doi.org/10.1534/g3.116.030411
Soto-Jimenez L, Estrada K, Sanchez-Flores A (2014) GARM: Genome Assembly, Reconciliation and Merging pipeline. Curr Top Med Chem 14:418–424. https://doi.org/10.2174/1568026613666131204110628
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/BIOINFORMATICS/BTU033
Stanke M, Diekhans M, Baertsch R, Haussler D (2008) Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24:637–644. https://doi.org/10.1093/bioinformatics/btn013
Vanhove M, Pina-Martins F, Coelho AC et al (2021) Using gradient forest to predict climate response and adaptation in cork oak. J Evol Biol 34:910–923. https://doi.org/10.1111/jeb.13765
Wu L, Nie L, Xu Z et al (2020) Comparative and phylogenetic analysis of the complete chloroplast genomes of three Paeonia section moutan species (Paeoniaceae). Front Genet 11:980. https://doi.org/10.3389/fgene.2020.00980
Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255
Yang Y, Zhou T, Duan D et al (2016) Comparative analysis of the complete chloroplast genomes of five quercus species. Front Plant Sci 7:959. https://doi.org/10.3389/fpls.2016.00959
Zoldos V, Papes D, Brown SC et al (1998) Genome size and base composition of seven Quercus species: inter- and intra-population variation. Genome 41:162–168. https://doi.org/10.1139/g98-006
Acknowledgements
The authors would like to thank Fundação João Lopes Fernandes for providing all plant materials.
Funding
Open access funding provided by FCT|FCCN (b-on). This research was funded by InAlentejo under the scope of “GenoSuber–Cork oak genome sequencing” (ALENT-07-0224-FEDER-001754), and by Alentejo2020, through FEDER under the scope “Lentidev-A genomic approach to cork quality” (ALT20-03-0145-FEDER-000020) and by Program PORTUGAL 2020 Partnership Agreement, under the scope of Biodata.pt–Infraestrutura Portuguesa de Dados Biológicos (22231/01/SAICT/2016), through the European Regional Development Fund (ERDF). Fundação para a Ciência e a Tecnologia (FCT), I.P., is acknowledged for funding researchers: Contrato–Programa to L. Marum (CEECINST/00131/2018), Contrato–Programa to A. Usié (CEECINST/00100/2021/CP2774/CT0001), and Research Contract to P. M. Barros (DL57/2016/CP1369/CT0029). O. Serra was funded by a Post-Doc fellowship under the research project “FASTBREED: implementation of a breeding program on wheat varieties based on genomic selection” (ALT20-03-0145-FEDER-000018). We also thank FCT for the financial support to Research Units UIDB/05183/2020 (MED-Mediterranean Institute for Agriculture, Environment and Development) and GREEN-IT-Bioresources for Sustainability (UIDB/04551/2020, UIDP/04551/2020) as well as LS4FUTURE (LA/P/0087/2020) Associated Laboratory.
Author information
Authors and Affiliations
Contributions
MMO, CPR, JM, FS, NS, CE, CMM, and SG conceived the study. SG, MMO, CPR, JM, and FS contributed to secure funding. FS, DM, JM, MCV, FN, ICa, PMB, MMO, ICh, CMM, SG, TC, and TA identified field material and collected biological samples. FS, PMB, DM JBG, LR, TC, and TA performed laboratorial experiments. AU and PB carried out the assembly, the removal of alternative heterozygous sequences, the assembly reconciliation of Illumina PE reads and the error correction of long PacBio reads and its de novo assembly. AU and OS performed all the scaffolding and gap filling procedures. AU annotated the genome with the involvement of CL who was responsible bioinformatics analysis of the RNA-Seq data. OS assembled and annotated the organelle genomes with the participation of AU. AU and PMB carried out the phylogenetic analysis. AU was responsible for the remaining bioinformatics analyses. AMR supervised the bioinformatics analyses. AU, OS, PMB, MMO, FS, and LM discussed and interpreted the results. AU, OS, and PMB wrote the manuscript. All the authors revised and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by M. Troggio
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 70 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Usié, A., Serra, O., Barros, P.M. et al. An improved reference genome and first organelle genomes of Quercus suber. Tree Genetics & Genomes 19, 54 (2023). https://doi.org/10.1007/s11295-023-01624-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-023-01624-8