Abstract
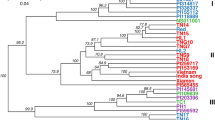
Since the start of avocado cultivation in South Africa, superior rootstocks and fruit cultivars have been selected based on morphological traits, which is time consuming and expensive. Technological advances, such as the development of a single nucleotide polymorphism (SNP) genotyping platform for avocado, may reduce these limitations. Therefore, the aim of this study was to implement molecular marker technologies for the validation of clonal material, verification of horticultural varieties, and determining the genetic diversity and population structure of an avocado cultivar germplasm in South Africa. An avocado cultivar breeding population, containing 375 individuals, was genotyped using 384 SNP markers. Our affinity propagation analysis (APA) indicated a 10.74% mislabelling in the germplasm. The principal component analysis (PCA) and discriminate analysis of principal components (DAPC) suggested that the germplasm was admixed in relation to the three known avocado varieties, Guatemalan, Mexican, and West Indian. Additionally, the ancestral origins were determined for 27 individuals with unknown ancestry. Furthermore, the population diversity was assessed and revealed moderate levels of differentiation in the germplasm, suggesting a high level of gene flow between the different populations. This research highlights the value of clonal verification and horticultural variety identification—for the reliable propagation of material with desired traits. The accurate propagation of material and clonal identity could aid avocado growers to link morphological characters and stress tolerance to accurate genetic backgrounds, which could improve the selection of avocados for current and future environmental stressors, especially as Africa is set to be significantly impacted by climate change.






Similar content being viewed by others
Data availability
The cultivar germplasm analysed during this study is available in the University of Pretoria Research Repository [Supplementary File 1—https://doi.org/10.25403/UPresearchdata.19145087].
References
Ashworth V, Clegg M (2003) Microsatellite markers in avocado (Persea americana Mill): genealogical relationships among cultivated avocado genotypes. J Hered 94:407–415
Batley J (2015) Plant genotyping. Humana Press, Dordrecht, London
Ben-Ya’acov A, Michelson E (1995) Avocado rootstocks. Vol. 17 (Janick J, ed.) John Wiley and Sons, Inc, New York, NY, pp 381–429
Bergh B, Ellstrand N (1986) Taxonomy of the avocado. Calif Avocado Soc Yearb 70:135–146
Bodenhofer U, Kothmeier A, Hochreiter S (2011) APCluster: an R package for affinity propagation clustering. Bioinformatics 27:2463–2464
Boza EJ, Tondo CL, Ledesma N, Campbell RJ, Bost J, Schnell RJ et al (2018) Genetic differentiation, races and interracial admixture in avocado (Persea americana Mill.), and Persea spp. evaluated using SSR markers. Genet Resour Crop Evol 65:1195–1215
Cañas-Gutiérrez GP, Arango-Isaza RE, Saldamando-Benjumea CI (2019) Microsatellites revealed genetic diversity and population structure in Colombian avocado (Persea americana Mill.) germplasm collection and its natural populations. J Plant Breed Crop Sci 11:106–119
Chen H, Morrell P, De La Cruz M, Clegg M (2008) Nucleotide diversity and linkage disequilibrium in wild avocado (Persea americana Mill.). J Hered 99:382–389
Chen H, Morell P, Ashworth V, De La Cruz M, Clegg M (2009) Tracing the geographic origins of major avocado cultivars. J Hered 100:56–65
Clegg M (2004) Application of molecular markers to avocado improvement. 24–28. California Avocado Commission, Proceedings of the California Avocado Research Symposium, University of California, Riverside. http://www.avocadosource.com/arac/symposium_2004/arac2004_pg_24.pdf. Accessed 21 May 2020
Crane J, Douhan G, Faber B, Arpaia M, Bender G, Balerdi C et al (2013) The avocado botany, production and uses: cultivars and rootstocks. In: Schaffer B, Wolstenholme B, and Whiley A (eds) CABI, pp 200–233
Davis J, Henderson D, Kobayashi M, Clegg M (1998) Genealogical relationships among cultivated avocado as revealed through RFLP analyses. J Hered 89:319–323
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Fisher RA (1936) The use of multiple measurements in taxonomic problems. Ann Eugen 7:179–188
Frey BJ, Dueck D (2007) Clustering by passing messages between data points. Science 315:972–976
Ge Y, Tan L, Wu B, Wang T, Zhang T, Chen H et al (2019a) Transcriptome sequencing of different avocado ecotypes: de novo transcriptome assembly, annotation, identification and validation of EST-SSR markers. Forests 10:411
Ge Y, Zhang T, Wu B, Tan L, Ma F, Zou M et al (2019b) Genome-wide assessment of avocado germplasm determined from specific length amplified fragment sequencing and transcriptomes: population structure, genetic diversity, identification, and application of race-specific markers. Genes 10:215
Gross-German E, Viruel M (2013) Molecular characterization of avocado germplasm with a new set of SSR and EST-SSR markers: genetic diversity, population structure, and identification of race-specific markers in a group of cultivated genotypes. Tree Genet Genomes 9:539–555
Guzmán LF, Machida-Hirano R, Borrayo E, Cortés-Cruz M, Espíndola-Barquera MdC, Heredia García E (2017) Genetic structure and selection of a core collection for long term conservation of avocado in Mexico. Front Plant Sci 8:243
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:1–15
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Juma I, Geleta M, Nyomora A, Saripella GV, Hovmalm HP, Carlsson AS et al (2020) Genetic diversity of avocado from the southern highlands of Tanzania as revealed by microsatellite markers. Hereditas 157:1–12
Karp A, Edwards KJ, Bruford M, Funk S, Vosman B, Morgante M et al (1997) Molecular technologies for biodiversity evaluation: opportunities and challenges. Nat Biotechnol 15:625–628
Köhne S (2005) Selection of avocado scions and breeding of rootstocks in South Africa. New Zealand and Australia Avocado Grower’s Conference, Tauranga. https://www.avocadosource.com/Journals/AUSNZ/AUSNZ_2005/KohneS2005.pdf. Accessed 30 Apr 2020
Kuhn D, Bally I, Dillon N, Innes D, Groh A, Rahaman J et al (2017) Genetic map of mango: a tool for mango breeding. Front Plant Sci 8:577
Kuhn D, Livingstone D III, Richards J, Manosalva P, van den Berg N, Chambers A (2019a) Application of genomic tools to avocado (Persea americana) breeding: SNP discovery for genotyping and germplasm characterization. Sci Hortic 246:1–11
Kuhn D, Dillon N, Bally I, Groh A, Rahaman J, Warschefsky M et al (2019b) Estimation of genetic diversity and relatedness in a mango germplasm collection using SNP markers and a simplified visual analysis method. Sci Hortic 252:156–168
Kuhn D, Groh A, Rahaman J, Freeman B, Arpaia M, van den Berg N et al (2019c) Creation of an avocado unambiguous genotype SNP database for germplasm curation and as an aid to breeders. Tree Genet Genomes 15:71
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Lachenbruch P, Goldstein M (1979) Discriminant analysis. Biometrics 35:69–85
Lahav E, Lavi U (2002) Genetics and classical breeding. In: Whiley A, Schaffer B, Wolstenholme B (eds.) The avocado: Botany, production and uses. CAB International, Wallingford, pp 39–69
Letunic I, Bork P (2019) Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256-259
Meirmans PG, Hedrick PW (2011) Assessing population structure: FST and related measures. Mol Ecol Resour 11:5–18
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci 70:3321–3323
Nei M, Chesser RK (1983) Estimation of fixation indices and gene diversities. Ann Hum Genet 47:253–259
Ottewell KM, Bickerton DC, Byrne M, Lowe AJ (2016) Bridging the gap: A genetic assessment framework for population-level threatened plant conservation prioritization and decision-making. Divers Distrib 22:174–188
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2:e190
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O et al (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830
Plotly Technologies Inc (2015) Collaborative data science. (Plotly Technologies Inc, ed.) Montréal, QC. https://chart-studio.plotly.com
Popenoe W, Williams L (1947) The expedition to Mexico of October 1947. Calif Avocado Soc Yearb 1947:22–28
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155
R Development Core Team (2020) R: a language and environment for statistical computing. R Foundation for Statistical Computing
Reich D, Price AL, Patterson N (2008) Principal component analysis of genetic data. Nat Genet 40:491–492
Rendón-Anaya M, Ibarra-Laclette E, Méndez-Bravo A, Lan T, Zheng C, Carretero-Paulet L et al (2019) The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc Natl Acad Sci 116:17081–17089
Rousseeuw P (1987) Silhouettes - a graphical aid to the interpretation and validation of cluster-analysis. J Comput Appl Math 20:53–65
RStudio Team (2016) RStudio: Integrated development for R. RStudio, PBC, Boston, MA. http://www.rstudio.com
Rubinstein M, Eshed R, Rozen A, Zviran T, Kuhn D, Irihimovitch V et al (2019) Genetic diversity of avocado (Persea americana Mill.) germplasm using pooled sequencing. BMC Genomics 20:379
Sánchez-González EI, Gutiérrez-Soto JG, Olivares-Sáenz E, Gutiérrez-Díez A, Barrientos-Priego AF, Ochoa-Ascencio S (2019) Screening progenies of Mexican race avocado genotypes for resistance to Phytophthora cinnamomi Rands. HortScience 54:809–813
Schaffer B, Wolstenholme B, Whiley A (2013) The avocado: botany, production and uses. CABI, Oxfordshire
Schleif R (1993) Genetics and molecular biology. Johns Hopkins University Press, Baltimore, MD
Schnell R, Brown J, Olano C, Power E, Krol C, Kuhn D et al (2003) Evaluation of avocado germplasm using microsatellite markers. J Am Soc Hortic Sci 128:881–889
Sneath PH, Sokal RR (1973) Numerical taxonomy. The principles and practice of numerical classification 1973. WH Freeman and Company, San Francisco
Williams L (1977) The avocado, a synopsis of the genus Persea, subg. Persea. Econ Bot 31:315–320
Winter DJ (2012) MMOD: an R library for the calculation of population differentiation statistics. Mol Ecol Resour 12:1158–1160
Wolstenholme B (2003) Avocado rootstocks: what do we know; are we doing enough research? South Afr Avocado Growers’ Assoc Yearb 26:106–112
Acknowledgements
The authors would like to thank the Forestry and Agricultural Biotechnology Institute (FABI) and the University of Pretoria for the use of their facilities and equipment. Furthermore, the authors would like to thank Dr David Kuhn for the custom affinity propagation scripts. Lastly, I would like to thank Allesbeste™ for providing the plant material.
Funding
The authors would like to thank the Hans Merensky Foundation© and Allesbeste™ for funding.
Author information
Authors and Affiliations
Contributions
RW contributed to the study design, experimental design, sample curation, formal analysis, investigation, visualisation, and drafting/writing/editing of the manuscript. NVDB contributed to the study conceptualisation and design, experimental design, project administration, resources, supervision, and funding. MMON and NA were responsible for methodology and technical assistance. PM provided the horticultural reference. BF extracted, processed, and performed the SNP genotyping. All co-authors contributed to writing/editing of the manuscript. All authors contributed to and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study received specific approval by the appropriate ethics committee for research involving plants.
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by C. Chen
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11295_2022_1573_MOESM1_ESM.pdf
Supplementary file1 Supplementary Fig 1 DAPC analysis of 159 genotypes indicating the cluster’s composition from K = 2 until K = 7 a) Bayesian information criterion (BIC) plot b) DAPC scatterplot - The eigenvalues and variance of each PC are found within parentheses on each axis. Individuals are represented as dots c) Genomic composition plot - Each thin vertical line in the bar plot represents one individual and each colour represents one inferred ancestral population. The length of each colour in a vertical bar represents the proportion of that individual’s ancestry that is derived from the inferred ancestral population corresponding to that colour. The same colour in different individuals indicates that they belong to the same cluster, indicating admixture (PDF 3451 KB)
11295_2022_1573_MOESM2_ESM.xlsx
Supplementary file2 Supplementary File 1: Formatted data of 326 genotyped individuals - Including industry suspected variety, DAPC assigned variety, assigned affinity propagation groups, silhouette scores, and genotype data (10.25403/UPresearchdata.19145087) (XLSX 663 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wienk, R., Mostert-O’Neill, M., Abeysekara, N. et al. Genetic diversity, population structure, and clonal verification in South African avocado cultivars using single nucleotide polymorphism (SNP) markers. Tree Genetics & Genomes 18, 41 (2022). https://doi.org/10.1007/s11295-022-01573-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-022-01573-8