Abstract
English walnut (Juglans regia L.) is an economically important crop with > 99% of US walnuts produced in California. Changes in climate and recent drought cycles have raised concerns regarding the future of nut production and responsible water use in California agriculture. Our study used an association genetics approach to characterize ecophysiological traits such as water use efficiency as estimated by carbon isotope discrimination (Δ13C), and photosynthetic capacity through foliar nitrogen composition, in important individuals of the Walnut Improvement Program, located at the University of California, Davis. Stable isotope and leaf measurements of 241 mature trees, representing 60 scion genotypes in established orchards were sampled in 2015 and 2016, followed by genotyping with the Walnut Axiom 700 k SNP Array. A mean Δ13C of 21.7‰ (σ: 0.9‰) was calculated for all individuals, as well as a mean nitrogen/leaf area (N/area) of 3.0 gN/m2 (σ: 0.5 gN/m2). A Bayesian analysis utilizing genomic relationships revealed rankings of the most water use efficient accessions as Solano (95-011-16), 67-013 (unreleased cross), and Tulare (67-011). 126,554 SNPs were used in a two-step association genetics approach identifying four loci associated with Δ13C after correction for multiple testing. Investigation of identified loci revealed an annotation on the J. regia genome of protein FAR1-related sequence 5-like, related to abiotic stress response. For uncharacterized markers, homologs were identified in Arabidopsis for two loci, similarly related to drought stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Persian, or English, walnut (Juglans regia L.) is the most economically important species of the Juglandaceae family, and is the only species of the Juglans genus widely cultivated for nut production (McGranahan and Leslie, 2012). In North America, walnuts are almost entirely grown in California, with a farm-gate value of $1.9 billion in 2014, and acreage has increased rapidly over the last decade to 127,500 bearing hectares (USDA NASS 2016). Walnut contributes substantially to California’s economy, with the only commodity walnut breeding program in North America located at the University of California, Davis. Established in 1948, the Walnut Improvement Program (WIP) has released all major cultivars grown in California, including the variety Chandler, which accounts for 53% of all bearing trees in California and 75% of all nursery sales (Ramos 1998; USDA-NASS 2016, 2017). While recent breeding in California has focused on production and yield-based traits, such as lateral bearing, harvest date, and kernel color (Martínez-García et al., 2017). However, few objectives in the program have addressed the environmental challenges facing California, such as water use.
Droughts in California are cyclical and are predicted to intensify as global climates shift, negatively impacting the agricultural water supply (Stine, 1994; Berg and Hall, 2017). Regardless, walnut acreage has continued to expand during one of California’s most severe multi-year droughts (Cooley et al. 2015; USDA NASS 2016). Water availability is crucial for proper development and production of walnuts. Furthermore, insufficient irrigation negatively impacts economic traits, such as kernel size and color (Ramos et al. 1978; Cohen et al. 1997). Historically, drought mitigation for walnut has been confined to orchard management practices. Unlike other nut crops, deficit irrigation techniques have not been as effective in maintaining yield while reducing water needs (Fulton and Buchner 2015). Incorporating water use efficiency as a breeding target has become an increasingly viable strategy in crop development for both cereal crops and legumes (Condon and Richards, 1992; Rebetzke et al. 2008; Dhanapal et al. 2015), but has remained a lower priority within the WIP in favor of yield and nut quality traits. This may be due in part to the limitations of perennial breeding programs (e.g., space, time, cost), but also to the complexity presented by plant water use and links to production.
Intrinsic water use efficiency, or the ratio of net carbon assimilation to stomatal conductance for water vapor, has been measured in a wide variety of species using carbon isotope discrimination (Δ) (Farquhar et al. 1989). The isotopic composition of carbon in leaf tissue is an established and inexpensive, high-throughput, indirect index of water use efficiency, making it ideal for phenotyping for large-scale breeding programs like WIP. The ratio of stable carbon isotopes varies both among and within species, and analyses have been carried out on several crop species, which show moderate to high broad-sense heritabilities, providing the potential for breeding gains (Impa et al. 2005; Rebetzke et al. 2008; Dhanapal et al. 2015). Studies in other tree species have also shown the potential of more efficient breeding using carbon isotope discrimination through associated markers (Cumbie et al. 2011; Marguerit et al. 2014; Guerra et al. 2016). Additionally, carbon isotope discrimination demonstrates substantial relationships with other growth factors such as leaf area, leaf thickness, and leaf nitrogen content, under well-watered conditions (Niinements, 1999; Prasolova et al. 2000; González-Martínez et al. 2008). Distinguishing the relationships of these additional traits is essential to elucidating water use efficiency and growth with regard to photosynthetic capacity or stomatal conductance and as a result of soil water availability (Handley et al. 1999; Niinements, 1999; González-Martínez et al. 2008, Cumbie et al. 2011; Easlon et al. 2013). Although carbon isotope discrimination is a known quantitative trait, the number of loci may not be conserved between species, making the underlying genetic architecture critical for breeding potential (McKay et al. 2003; Casasoli et al. 2006; Collard and Mackill, 2008; Tsai et al. 2015). WIP utilizes classical, phenotypic, breeding approaches for current target traits, but recent developments in walnut genomics have increased opportunities for targeted genetic improvement in walnut breeding (Bernard et al. 2018). The availability of a walnut reference genome and the subsequent Axiom 700 k SNP array have created new opportunities for including more complex traits using genomic breeding methodologies (Martínez-García et al. 2016; Marrano et al. 2018).
For this pilot study, our objectives were to characterize phenotypic variation and calculate estimated breeding values of founding and significant cultivars in the WIP for the following traits: carbon isotope discrimination (Δ13C), leaf carbon and nitrogen content and ratio, leaf nitrogen isotope composition, as well as, specific leaf area (SLA) and nitrogen content per leaf area (N/area). An additional objective was to use the newly developed Walnut SNP 700 k SNP Array (Marrano et al. 2018) to perform genome-wide association studies to identify possible makers for future breeding purposes.
Materials and methods
Germplasm material and growth conditions
Leaves were collected in September of 2015 and 2016, from the upper crown of 241 mature trees (60 unique genotypes), growing across five established WIP orchards with 20′ to 30′ spacing, located at the University of California, Davis (38° 32′ 22.7″ N, 121° 47′ 44.2″ W). Genotypes reflected five generations of germplasm material, represented by between two to eight clonal individuals per genotype. Genotypes were selected for their prevalence in the historical pedigree, current production, and future breeding design of walnut cultivars in California. Orchards were fertilized annually with standard UN-32 (45% ammonium nitrate, 35% urea, 20% water) every 4 months, with total applications between 120 and 140 lbs./acre. Additionally, orchards were regularly irrigated to avoid water deficits, though water retention may have differed among individual plots due to variable clay soil composition.
Prior to 2005, clonal rootstocks were not utilized in the WIP germplasm blocks; therefore, genetic scion-rootstock combinations were not replicated in this experiment. Although genetically unreplicated within our experiment, all rootstocks used in this study are industry standards for California growers, representing actual production conditions of the walnut industry.
Phenotypic measurements
Two primary leaflets were collected from four of the youngest, fully expanded, leaves from the southeast side of each tree crown, for a total of eight leaflets per tree. Each leaf was imaged using a Canon PowerShot ELPH 100HS digital camera (Canon Inc., Tokyo, Japan). Images were analyzed using Easy Leaf Area V2 (Easlon and Bloom 2014) to determine an average measurement of individual leaf area per tree. Following these measurements, midribs were removed, leaves were dried at 45 °C for 48 h, and leaves were weighed to the nearest hundredth gram. Per tree individual leaf area and weight averages were used to calculate the specific leaf area (SLA) of each tree with the following equation:
Leaves were then pooled and ground to a fine powder using a mini-beadbeater-24 (BioSpec, Bartlesville, OK) and submitted to the stable isotope facility (SIF, University of California, Davis, CA) following submission guidelines.
Isotopic analyses of 13C and 15N were measured on two aliquots of each sample, using a continuous flow isotope ratio mass spectrometer (IRMS) located at SIF. Composition values were reported relative to international standards (Vienna PeeDee Belemnite for C, and air for N) using the following equation:
Where δp, the composition of the plant tissue, is a function of the isotopic abundance of the plant material (Rp) and the standard (Rs) (Farquhar et al. 1989). Raw 15N composition values were used for further analysis, while carbon isotope discrimination values (Δ) were calculated relative to the 13C composition of air, using the following equation:
Where Δ is the change in 13C composition from the composition of 13C in air (δa), assumed to be −8‰, to the 13C composition of the plant (δp).
Additionally, a subset of 36 individuals of as many cultivar genotypes was selected for estimation of photosynthetic capacity. Genotypes used in this subset were selected as a representation of the phenotypic diversity of both %N content and Δ13C, and were measured with a portable photosynthesis system, LiCor 6400 (LI-COR, Lincoln, NE). Measurements of maximum net assimilation (Amax), stomatal conductance (gs) and internal carbon concentration (Ci) were recorded for three leaves in the lower southeast canopy at both “normal” CO2 concentration (400 ppm), and “saturated” concentration (1500 ppm). All traits previously mentioned were repeated for this subset following gas exchange measurements.
Genotypic data
DNA was extracted from the leaf tissue of one individual of each cultivar using the E.Z.N.A plant DNA kit (Omega Bio-Tek, NorCross, GA) and sent for single nucleotide polymorphism (SNP) genotyping on the Walnut Axiom 700 k SNP array (Marrano et al. 2018; Affymetrix, Santa Clara, CA). SNPs were identified at both the inter- and intra-species level, from the resequencing of 4 Juglans species (Stevens et al. 2018) and a panel of 27 founding individuals of the WIP (Marrano et al. 2018). Polymorphisms were filtered as described by Marrano et al. (2018) resulting in a 609,658 SNP array, spanning 627 Mbp of the genome. Genotyping results for all 60 genotypes were filtered for conversion quality using the Axiom Suite software resulting in 146,182 markers. Additional filtering using the public domain software GenABEL (Aulchenko et al., 2007) accessed through R (R Core Team, 2017) for call rate (> 95%), and minor allele frequency (MAF > 5%), reduced the number of high quality SNPs to 126,654 SNPs for association analysis. Variants included on the SNP array focused on the inclusion of markers within genic regions, and even distribution throughout the genome (Marrano et al. 2018).
Trait analyses
To elucidate unrecorded relationships of WIP germplasm, both historical and genotypic data were used to construct kinship matrices using the open-source software Synbreed (Wimmer et al. 2012) through the public-domain language R (R Core Team, 2017). The pedigree-based matrix was constructed from known ancestry of the 60 unique cultivars within WIP while a similar marker-based kinship matrix was created by averaging 100 iterations of 20 k randomly selected markers from the 126,654 available SNPs.
To assess the impact of genotype on our assembled phenotypes, we utilized a series of commonly considered mixed linear models. Our objective was to simultaneously evaluate differences between cultivars, estimate heritability and predict breeding values for each cultivar represented in the WIP. All analyses conducted, including several preliminary analyses used to assess important explanatory variables, are extensions of the general mixed linear model. Importantly, however, these models reflect that several clones were evaluated within each cultivar, and that within each clone, laboratory analyses were repeated to enhance the accuracy of phenotyping. Accordingly, our models include terms for a repeated measures analysis, permitting the simultaneous estimation of repeatability, along with the heritability of each trait.
The algebraic form for our model is
where yijk is the k-th (k = 1,2) observation on the j-th clone (j = 1, 2,…, ni, with the total number of clones being 241) within the i-th cultivar (i = 1, 2,…, 60), μ is a constant common to all phenotypes, cultivari is the random effect of each cultivar representing the additive genetic merit of each cultivar, cloneij is the random contribution of clones within each cultivar representing the repeated sampling of these phenotypes within a cultivar and eijk is the random residual associated with the ijk-th phenotype. The three random effects are assumed to be normally distributed, independent of one another and to have null means. Using matrix notation to denote the vectors of cultivar, clone, and residual effects as a, p, and e, respectively, we assume the covariance structure to be
where \( {\sigma}_a^2 \), \( {\sigma}_p^2 \), and \( {\sigma}_e^2 \) are unknown variances for the additive genetic values, the permanent environmental effects and residuals, respectively, K is a known genotypic covariance matrix of order 60 for the cultivar effects, with I as an identity matrix of order 241 for the permanent environmental terms represented in the clones and I is an identity matrix of order N for the residuals. Therefore, the heritability of each trait can be evaluated from the ratio \( {h}^2={\sigma}_a^2/\left({\sigma}_a^2+{\sigma}_p^2+{\sigma}_e^2\right) \) and the repeatability of each trait (the correlation of repeated observations on the same cultivar) from the ratio \( r=\left({\sigma}_a^2+{\sigma}_p^2\right)/\left({\sigma}_a^2+{\sigma}_p^2+{\sigma}_e^2\right) \).
This analysis simultaneously estimates the unknown constant μ, predicts cultivar breeding values in a and permanent environmental contributions of clones in p, along with providing estimates of the unknown variances, \( {\sigma}_a^2 \), \( {\sigma}_p^2 \), and \( {\sigma}_e^2 \). Evaluating these parameters was facilitated through a hierarchical Bayesian framework implemented through the public-domain software Stan (Carpenter et al. 2016), accessed through R (R Core Team, 2017).
To utilize this Bayesian framework, prior densities must be established for unknown model parameters. Specifically, we must assume a diffuse normal prior for the unknown constant (i.e., a normal density with null mean and standard deviation 103) along with normal densities (as outlined above) for the cultivar, clone, and residuals (denoted as a, p, and e). For the unknown variance components, \( {\sigma}_a^2 \), \( {\sigma}_p^2 \), and \( {\sigma}_e^2 \), we chose the weakly informative prior of a half-Cauchy (0,5), where samples were limited to positive values for each parameter (Gelman 2006). In order to evaluate this hierarchical model, we utilized a Monte Carlo Markov Chain (MCMC) sampling process run in four chains. Each chain was based upon 15,000 total samples with the first 5000 discarded as part of the warm-up process and the remainder thinned to every 20th sample, resulting in a MCMC sample of 2000 values (Carpenter et al. 2016). The convergence to the posterior density was evaluated by the Gelman-Rubin test statistic and values below 1.05 indicate the adequacy of the MCMC sample process for the data evaluated (Gelman and Rubin 1992).
Association analyses
All association testing was done using a two-step approach in the public-domain software GenABEL (Aluchenko et al. 2007) accessed through R (R Core Team, 2017). A similar genomic relationship matrix, calculated from 20 k SNPs within the GenABEL package, was used to address underlying bias in the association model and tested using a FAmily-based Score Test of Association (FASTA) approach (Chen and Abecasis, 2007). Initial estimations of the variance-covariance matrix and trait residuals were calculated using the polygenic function and subsequently used for association testing with the function mm score (Thompson and Shaw, 1990; Chen and Abecasis, 2007). For each trait, a genomic deflation factor (λ) was calculated and used as a correction factor. λ ranged from 0.99 for Δ13C to 0.87 for both %C and N/area.
The P values observed as a result of the association models were compared to a uniform distribution of expected P values. Using the mean of squared differences between observed and expected P values, the strength of deviation was calculated to determine the importance of individual markers (Stich et al. 2008). A modified Bonferroni correction was used to calculate a significance threshold for multiple testing of individual markers using the calculation \( \frac{1}{n\ SNPs} \). Results were visualized using the R package QQ-Man (Turner 2014).
SNP annotation
For each trait, SNPs with the greatest association (highest –log10 P values) were compared to known annotations of the Juglans regia V1.0 genome (taxid: 51240; Martínez-García et al. 2016). Upstream and downstream probe sequences (71 bp) were used in a BLASTx megablast query of the NCBI refseq protein database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Parameters such as “expect threshold” and “low complexity filters” were broadened to increase possible annotations from the reference genome. Annotations of associated SNPs were reported, along with chromosome, P values, and minor allele frequencies. Uncharacterized locations of significant SNPs were further analyzed using TAIR BLAST 2.2.8 (Lamesch et al. 2012) for possible comparative Arabidopsis annotations.
Results
Population relationships and trait analysis
Following the application of standard filters in GenABEL, 126,654 SNPs were retained for association analysis. The majority of SNPs (82.7%) had a minor allele frequency (MAF) > 0.1. The mean heterozygosity for each SNP was 0.33 (sd 0.13), and heterozygosity of individual cultivars had a mean of 34% (sd 5.4%) for all loci. Principal component analysis revealed no discernable population substructure or clustering, with the first three components accounting for ~ 40% of the variation (Fig. S1).
Two relationship matrices were constructed to elucidate relationships between cultivars (Fig. 1). By using historical records, the average kinship coefficient was 0.12. Additionally, minor amounts of inbreeding were identified in the youngest generation as expected given the visualized pedigree (Fig. S2), though substantial gaps of information (represented by a lack of color in relationship matrices) remain for the founders of the breeding program when relying on historical records (Fig. 1a). The same relationships were then estimated using the marker information resulting in higher estimations of inbreeding and close relationships between cultivars with no pedigree information (Fig. 1b). The realized relationship matrix identified an average kinship coefficient of 0.78, and identified three lesser-related individuals within the program, PI159568 (48), Manregian (49), and Xinjiang 6 (85-008).
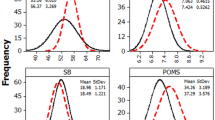
Estimated kinship values were incorporated into a mixed model analysis with a hierarchical Bayesian approach, to estimate trait heritabilities and cultivar trait rankings. For Δ13C, convergence for both heritability and repeatability appeared consistent across all four chains with uni-modal density distributions. Of the 2000 sampled values, the effective N (Neff) for heritability was 2000 for the pedigree-based analysis and 1978 for the same analysis utilizing the genomic relationships (Table 1). The lowest Neff was for the pedigree-based analysis of the C/N ratio, with 1243 sampled values. Convergence for other traits was similarly stable among the four chains and density distributions for both heritability and repeatability.
Estimations of narrow-sense heritability (h2) increased for all traits when using the marker-based relationships as opposed to the pedigree-based approach (Table 1). However, most traits maintained low narrow-sense heritabilities. Leaf carbon to nitrogen ratio had one of the highest h2 of 0.17 (CI: 0.13, 0.21) using the realized matrix, followed by Δ13C (h2 = 0.14; CI: 0.11, 0.17), while N/area remained the least heritable of the traits with a h2 of essentially 0 (Table 1). Analysis of SLA required adjustments to priors resulting in very high estimates of heritability (h2 = 0.94; CI: 0.91, 0.97) and repeatability (r2 = 0.95; CI: 0.92, 0.98). Further investigation of this trait is necessary to determine the true underlying genetic relationship. Although these results suggest SLA is a predominantly genetic trait, with little environmental contribution, previous studies have reported high phenotypic plasticity of SLA, primarily affected by nutrient availability (Braatne et al. 1992) as well as light exposure (sun versus shade leaves) (Lambers et al. 2008).
Predicted cultivar effects estimated from the pedigree information, estimated breeding values (EBVs) were compared to effects estimated from the realized matrix, genomic estimated breeding values (GEBVs) (Table S1). GEBVs showed increased variation as compared to pedigree based EBVs for Δ13C (Fig. 2, Table S1). Additionally, other traits saw increases in breeding value variation, though some ranges remained consistent (Table S1). GEBVs were then used to rank cultivars for trait performance for future breeding potential in addition to the estimated breeding values using the pedigree. (Table S2). The top three cultivars with breeding potential for the lowest discrimination, and therefore highest breeding potential for water use efficiency, were Solano (95-11-16), 67-013 (unreleased cross), and Tulare (67-011), respectively (Table S2). Cultivars with the greatest Δ13C (discrimination, or least efficient) breeding potential (03-001-2434, 03-001-977, and 91-096-3) are all unreleased crosses under evaluation for other traits. In addition, the cultivar Chandler (64-172), a high yielding cultivar and frequently used crossing parent, maintained a lower ranking for Δ13C at 56th, and 52nd for %N, yet a higher ranking, 10th, for the C/N ratio (Table S2).
Correlations of GEBVs showed that many calculated breeding values were not significantly correlated. However, area-related traits, which were not phenotypically correlated to Δ13C (see below), demonstrated significant genetic relationships. Δ13C presented a positive correlation of r = 0.46 with SLA (P value = 0.0002), and a significantly negative correlation of r = −0.42 with N/area (P value = 0.0008) (Table 2). Conversely, N/area presented a significantly positive correlation with δ15N (r = 0.48, P value = 0.0001). %C and %N were negatively genetically correlated (r = −0.32, P value = 0.014) which were consistent with the phenotypic correlation (see below).
Trait summaries and phenotypic variation
Measurements of carbon discrimination (Δ13C), nitrogen composition (δ15N), percent content and ratio of carbon and nitrogen (%C, %N, and C/N, respectively), as well as specific leaf area (SLA) and nitrogen per unit leaf area (N/area), were measured for all 241 individuals of 60 unique genotypes. Between individuals, values of leaf Δ13C ranged from 19.3‰ to 24.4‰, with a mean of 21.7‰, displaying significant phenotypic variation within the population (Table 3). More than twofold variation was seen in nitrogen-related traits: %N, C/N, and N/area. For δ15N, the mean was 4.2%, and had a large range of 1.6% to 8.3% (Table 3). SLA also showed substantial variation among genotypes, with a mean of 99.1 m2/kg leaf (sd. 20.2 m2/kg), and a maximum of 203.2 m2/kg (Table 3).
Significant variation was observed between genotypes for all traits; however, initial analysis showed significant variation within genotypes as well. Due to the unbalanced nature of established orchards, further explanation of within genotype variation remains undetermined from this pilot study. Effects of rootstock, age and location were confounded within this pilot study and could not be differentiated from the residual error term.
Correlations between phenotypic traits showed predominantly negative relationships. Pearson’s correlation coefficients were calculated for all independent traits of both isotope and leaf measurements. Correlations ranged from −0.48 for C/N and SLA, to 0.46 for %N and SLA (Table 4). Other high negative correlations were seen between %C and %N, and %C with SLA, with correlations of r = −0.38 and r = −0.27 respectively (Table 4).
Stable isotope measurements collected in 2015 were compared to the isotope measurements of the subset of leaves collected in 2016, and showed high positive correlations among nitrogen related traits (%N: r = 0.65; C/N: r = 0.60; δ15N: r = 0.75; all P values < 0.001). Additionally, significant correlations were seen between Amax at 1500 ppm CO2 for %N (r = 0.45, P value = 3.2E−05), and N/area (r = 0.44, P value = 4.19E−05). However, no significant correlations arose between Δ13C and any of the LiCor measurements: Amax, gs, A/gs, or Ci. Furthermore, the addition of %N as a covariate had no significant effect on the correlation of Δ13C with Amax at 1500 ppm CO2.
Marker-trait association analysis and SNP annotations
Following correction for multiple testing, association analysis revealed 4 SNPs observed to be above expectations for Δ13C (Fig. 3a). Significantly associated SNPs for Δ13C were located across three chromosomes 1, 2, and 16 (Fig. 3b), with the most highly associated location in chromosome 1 (P value = 3.07E−07). Correlation investigation of the two markers on chromosome 16 showed no significant linkage disequilibrium (LD) (r = 0.00043). While other traits did not see markers above expectation, QQ plots of these traits showed trends consistent with similar analyses in other species (Fig. S3). Furthermore, despite having no individual SNP associations above the significance threshold, signal patterns of association on similar chromosomes were present (Fig. S4).
Investigation of the top 100 SNPs showed substantial overlap of markers between traits, the highest being %C and C/N overlapping 97%, though this is likely attributed to the dependent nature of these two traits (Table 5). A majority of SNPs were contained in chromosome 1 for %C (and therefore C/N) as well as for δ15N, and N/area, which is consistent, given the genetic correlations from GEBVs. Top SNPs associated with %N differed slightly with a quarter of SNPs coming from chromosomes 11 and 13, while half of the top SNPs for SLA were primarily located on chromosomes 3 and 13. Of the 700 selected, 416 were unique, with N/area showing an overall high incidence of overlap with other traits (Table 5). Additionally, genetic correlation of GEBVs, SLA with δ15N was positive, yet only 3% of top SNPs overlapped (Tables 2, 5). Conversely, half of the top SNPs for δ15N were shared with %C, yet GEBV correlation was not significant. Similarly, δ15N and C/N ratio showed 50% overlap with top SNPs while a strong negative genetic correlation was seen (Tables 2, 5). One SNP, 171499827, showed overlap with six of seven traits (Table S3).
Annotations were investigated for the four significant SNPs associated with Δ13C using BLASTx queries. Annotation for the one characterized Δ13C SNP within J. regia was an uncharacterized aarF domain containing protein kinase, At1g71810 chloroplastic-like (E value = 1.9). Additional annotations were performed using the TAIR database and resulted in potential annotations for all four associated SNPs, including an early response dehydration stress protein (ERD4) for the most significant SNP (Table 6). In addition to the four associated SNPs for Δ13C, the 416 unique SNP sequences were used in BLASTx queries resulting in 197 SNPs providing 254 annotations (Table S3). Of these annotated SNPs, the minor allele frequency (MAF) ranged from 0.05 to 0.50, with an average MAF of 0.24 (Table S3). The most common annotations of the characterized SNPs involved regions of serine/threonine kinase activity and zinc finger domain containing proteins (Table S3). Additionally, some annotations of lectin properties for δ15N were similar in other tree species, Pinus taeda and Populus trichocarpa (Cumbie et al. 2011; Guerra et al. 2016) (Table S3).
Discussion
As the strain on California’s agricultural water supply continues, targeted breeding for water use efficiency in perennial crops, such as walnut, will rise in priority (Condon et al. 2004; Aletà et al. 2009; Bassett et al. 2011). Demand for agricultural water will persist in walnut production as production acreage and crop exportation increases. While walnut is known to utilize stomatal regulation during dry periods as a mechanism of “drought avoidance,” impacts on kernel quality are evident when water is limited (Cohen et al. 1997; Rosati et al. 2006). Assessment of germplasm under well-watered conditions was used to avoid damage of the WIP germplasm collection as previous studies have shown that walnut is highly responsive to environmental stresses with sensitive stomata and susceptibility to cavitation and embolism (Cochard et al. 2002; Rosati et al. 2006).
Careful management of water resources will be essential for the future of agriculture; targeting water-use efficiency in breeding programs while maintaining yield is critical in the face of a changing climate (Condon et al. 2004; Cattivelli et al. 2008). However, breeding in horticultural crops can be significantly complicated by long-generation times and high cost (van Nocker and Gardiner, 2014). Long juvenile phases increase the time between selection and crosses by delaying proper phenotypic assessment. Additionally, the size of individual trees places a costly strain on the number of replicates making marker-assisted or genomic selection an attractive strategy (Goddard and Hayes, 2007; van Nocker and Gardiner, 2014). Marker identification through association studies and prediction models require robust measurements and specifically selected individuals in order to adequately estimate heritability in a specific population (Visscher et al. 2008).
Plant responses to water deficit are complex and water loss strategies vary between plant species. Previous studies have shown that Juglans regia displays phenotypic and genetic diversity for water use efficiency (WUE) based on origin, though, to date, no studies have characterized heritability or potential markers for use in walnut breeding (Aletà et al. 2009). Carbon isotope discrimination (Δ13C) is a known method to estimate or index WUE within species and has moderate genetic control (Farquhar et al. 1989; Condon et al. 2004). The identification of genetic markers which associate with Δ13C and markers associating with other growth traits, such as specific leaf area (SLA) and nitrogen metabolism (%N, δ15N), are valuable to walnut production and may elucidate similar genetic loci in other species (Handley et al. 1999). Additionally, a better understanding of the genetic diversity within WIP will benefit future breeding strategies.
Through our pilot study, the phenotypic diversity of Δ13C and water-use efficiency traits was characterized within WIP for future breeding strategies. Additionally, these relationships were compared with a subset of individuals for gas exchange with a LiCor 6400. Expected relationships were unclear in the role of Δ13C as an estimation of photosynthetic capacity or stomatal conductance, and may have been the result of canopy variation possibly and carbon assimilation within tree crowns as seen previously by Le Roux et al. (1999).
Though correlations were not observed between 2015 and 2016 isotope measurements for carbon traits, nitrogen-related traits showed significant correlations implying genetic control in these growth traits. Additionally, positive correlations for both %N and N/area with photosynthetic rate were consistent with previous literature (Hikosaka and Terashima, 1996, Niinements, 1999). While Δ13C values did not significantly correlate with Amax, nor Amax/gs, measurements, the trend of a negative relationship was observed providing justification for further investigation of Δ13C with the possibility of breeding potential (Brendel et al. 2008). Additionally, the negative relationship Δ13C and %C coupled with the positive correlation of %C and %N could elucidate the positive correlation of %N and Amax suggesting that variation of Δ13C could be more strongly controlled by stomatal conductance than photosynthetic capacity as previous studies have shown (Rosati et al. 2006; Aletà et al. 2009). Broader experiments within the breeding program should be considered for future field measurements to identify connections between mesophyll conductance and the effects of evaporative demand on water-use efficiency (Condon et al. 2004; Seibt et al. 2008).
Analysis of kinship within the WIP revealed significant inbreeding and rapid potential for significant loss of diversity within the program. Historical records showed that many previous crosses made from European cultivars, may already have been bred within a highly related lineage (Tulecke and McGranahan 1994). Pedigree-based relationships alone denote inbreeding in some of the youngest generation with coefficients as high as 1.16. These founder relationships were further explained through the marker-based matrix showing a definite increase in relatedness among individuals that were previously characterized as unrelated. By estimating relationships based on the amount of homozygosity at loci, we assume that these genotypes are the result of common ancestry and thus increase the kinship coefficients (Bernardo et al. 1996; Powell et al. 2010). While inaccurate relationships can be reported from these analyses, we have accounted for marker bias by including several genotypes used in the design of the Walnut 700 k SNP Array in our population (Powell et al. 2010). The average relationship within the marker-based matrix rose to 0.78, leaving only three individuals displaying more distant relationships. This estimated matrix confirms the historical mating design through these three less-related individuals. Xinjiang 6 (85-008) was added to the program in 1985 following a collection of international germplasm. Additionally, PI159568 (48; added in the 1930s) and Manregian (49; added in the early 1900s) are known to be founders in the breeding program.
Using the relationship matrices heritability and genomic estimated breeding values (GEBVs) were calculated with GEBVs being utilized for genotype breeding rankings (Table 3; Table S2). While heritabilities remained low, similar narrow-sense heritabilities (low to moderate) have been reported for Δ13C in other species, e.g., Pinus taeda (Baltunis et al. 2008); Pinus pinaster (Marguerit et al., 2014); and Quercus robur (Brendel et al. 2008). Additionally, low h2 may be indicative of a more complex or polygenic trait, similar to other reported studies of these traits (Cumbie et al. 2011; Guerra et al. 2016). While complex traits are inherently more difficult to predict and control in traditional breeding programs (Hill et al. 2008), especially when coupled with low heritability, labor-intensive phenotypic measurements can inflate the cost of the selection process making them prime targets for genomic selection models (Goddard and Hayes, 2007).
GEBV rankings for our analyzed traits provide a first look at the future of water efficiency and walnut production. Expectedly, Chandler, which makes up three quarters of the current production acreage due to its high yield and extra light kernel color, ranked among the lowest for WUE (56th). This is similar to high-yielding varieties in other crops where WUE has been unconsciously selected against by way of increased production inputs (Condon et al. 2004). Interestingly, the highest ranked genotypes for WUE include two more recent cultivar releases, Solano (95-016-11) and Tulare (67-011), and are both considered high-yielding with desirable kernel traits. Further consideration of these cultivars for crosses should involve increased characterization of stress-related efficiency traits in addition to current trait evaluations.
Correlations of these GEBVs revealed significant relationships between Δ13C and leaf area, demonstrated by the simultaneous positive relationship with SLA and negative relationship with N/area. These inverted correlations of 13C accumulation with different area traits could be the result of a stronger role of stomatal conductance in carbon assimilation rather than a limitation of photosynthetic capacity (Rosati et al. 2006). Additionally, the lack of significant GEBV correlations between Δ13C and nitrogen traits supports the findings of others that these may be independent genetic relationships, providing the possibility of breeding for WUE while maintaining tree growth (Marguerit et al., 2014). However, further examination of growth under stress should examine the positive phenotypic relationship of Δ13C with δ15N, an indicator of nitrogen accumulation under different soils moistures, implying that WUE could come at the cost of production (Handley et al. 1999, Condon et al. 2004).
Markers exceeding the correction threshold were identified for Δ13C, while other traits showed signals in consistent chromosomes. SNPs associated with Δ13C were found in uncharacterized regions with two providing only one annotation on the J. regia genome providing possible functional annotation (Table 6). A single annotation to the J. regia genome, an uncharacterized aarF domain containing protein kinase revealed a TAIR annotation of ArfGap/RecO-like zinc finger domain containing proteins which have been seen to be involved in changes in expression, some specifically relating to abiotic stress (Laity et al. 2001; Wang et al. 2008). Additional annotations revealed from the TAIR database identified other possible abiotic stress response genes such as early response dehydration stress protein (ERD4) involved in drought recovery and regulated by abscisic acid (ABA) production (Huang et al. 2008), clathrin adaptor complexes, and ARM repeat super family protein.
While our analysis revealed no significant markers for six of the seven traits examined, the top 100 markers were used in annotation analysis. An investigation of top markers revealed annotations consistent with that of other investigated tree species (Gonzalez-Martinez et al. 2008; Cumbie et al. 2011; Guerra et al. 2016). The most common characterizations of SNP sequences were located in regions of serine/threonine kinase activity and metal ion binding, specifically zinc finger domain containing proteins, previously mentioned to have possible involvement in drought stress responses (Wang et al. 2008). Although many traits shared markers, and these are confined to only a few chromosomes, these loci should be further investigated before conclusions can be drawn regarding their relationships.
Conclusion
The results of our preliminary study provide a basis for the potential of gains within the walnut breeding program. While no single signals were seen, which would be indicative of major gene responses, many loci appear to be controlling specific growth and efficiency traits, such as Δ13C, and %N. It is likely that each of these loci produce small effects consistent with the quantitative genetics theory of the infinitesimal model (Fisher, 1918; Falconer and MacKay, 1996). Additionally, these traits demonstrate a low but highly repeatable narrow-sense heritability within WIP justifying future research opportunities and potential for genomic selection. As the breeding program expands, increased phenotyping will be necessary to assess the full potential of the program and maintain accurate estimates of heritability along with well-trained selection models.
Further analysis of these SNPs within the greater breeding population will identify allele frequencies and individual SNP effects within the program. Additionally, new genetic maps may further characterize SNP regions, as our markers were generally limited to a small number of chromosomes. Likewise, measurement and analysis of large families utilizing seedling, or grafted individuals within the program may more finely identify associated regions within the genome and investigate the interaction of rootstock and scions in water use efficiency. The results of this study provide opportunities for water-use efficient breeding in the Walnut Improvement Program at UC Davis.
References
Aletà N, Vilanova A, Díaz R, Voltas J (2009) Genetic variation for carbon isotope composition in Juglans regia L.: relationships with growth, phenology and climate of origin. Ann For Sci 66(4):413
Aulchenko YS, Ripke S, Isaacs A, Van Duijn CM (2007) GenABEL: an R library for genome-wide association analysis. Bioinformatics 23(10):1294–1296. https://doi.org/10.1093/bioinformatics/btm108
Baltunis BS, Martin TA, Huber DA, Davis JM (2008) Inheritance of foliar stable carbon isotope discrimination and third-year height in Pinus taeda clones on contrasting sites in Florida and Georgia. Tree Genet Genomes 4(4):797–807. https://doi.org/10.1007/s11295-008-0152-2
Bassett CL, Glenn DM, Forsline PL, Wisniewski ME, Farrell RE (2011) Characterizing water use efficiency and water deficit responses in apple (Malus X domestica Borkh. and Malus sieversii Ledeb.) M. Roem. Hort Sci 46(8):1079–1084
Berg N, Hall A (2017) Anthropogenic warming impacts on California snowpack during drought. Geophys Res Lett 44(5):2511–2518. https://doi.org/10.1002/2016GL072104
Bernard A, Lheureux F, Dirlewanger E (2018) Walnut: past and future of genetic improvement. Tree Genet Genomes 14(1):1. https://doi.org/10.1007/s1129
Bernardo R, Murigneux A, Karaman Z (1996) Marker-based estimates of identity by descent and alikeness in state among maize inbreds. Theor Appl Genet 93(1–2):262–267. https://doi.org/10.1007/Bf00225755
Braatne JH, Hinckley TM, Stettler RF (1992) Influence of soil-water on the physiological and morphological components of plant water-balance in Populus trichocarpa, Populus deltoides and their F1 hybrids. Tree Physiol 11(4):325–339
Brendel O, Thiec DL, Scotti-Saintagne C, Bodénès C, Kremer A, Guehl JM (2008) Quantitative trait loci controlling water use efficiency and related traits in Quercus robur L. Tree Genet Genomes 4(2):263–278
Carpenter B, Gelman A, Hoffman M, Lee D, Goodrich B, Betancourt M, Brubaker M, Riddell A (2016) Stan: a probabilistic programming language. J Stat Softw 20:1–37
Casasoli M, Derory J, Morera-Dutrey C, Brendel O, Porth I, Guehl JM, Villani F, Kremer A (2006) Comparison of quantitative trait loci for adaptive traits between oak and chestnut based on an expressed sequence tag consensus map. Genetics 172(1):533–546. https://doi.org/10.1534/genetics.105.048439
Cattivelli L, Rizza F, Badeck FW, Mazzucotelli E, Mastrangelo AM, Francia E, Mare C, Tondelli A, Stanca AM (2008) Drought tolerance improvement in crop plants: an integrated view from breeding to genomics. Field Crop Res 105(1–2):1–14. https://doi.org/10.1016/j.fcr.2007.07.004
Chen WM, Abecasis GR (2007) Family-based association tests for genomewide association scans. Am J Hum Genet 81(5):913–926. https://doi.org/10.1086/521580
Cochard H, Coll L, Le Roux X, Ameglio T (2002) Unraveling the effects of plant hydraulics on stomatal closure during water stress in walnut. Plant Physiol 128(1):282–290. https://doi.org/10.1104/Pp.010400
Cohen M, Ameglio T, Cruiziat P, Archer P, Valancogne C, Dayau S (1997) Yield and physiological responses of walnut trees in semiarid conditions: application to irrigation scheduling. Acta Hortic 449:273–280
Collard BCY, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc., B 363(1491):557–572. https://doi.org/10.1098/rstb.2007.2170
Condon AG, Richards RA (1992) Broad sense heritability and genotype x environment interaction for carbon isotope discrimination in field-grown wheat. Aust J Agric Res 43(5):921–934. https://doi.org/10.1071/Ar9920921
Condon AG, Richards RA, Rebetzke GJ, Farquhar GD (2004) Breeding for high water-use efficiency. J Exp Bot 55(407):2447–2460. https://doi.org/10.1093/jxb/erh277
Cooley H, Donnelly K, Phurisamban R, Subramanian M (2015) Impacts of California’s ongoing drought: agriculture. Pacific Institute, Oakland, p 24
Cumbie W, Eckert A, Wegrzyn J, Whetten R, Neale D, Goldfarb B (2011) Association genetics of carbon isotope discrimination, height and foliar nitrogen in a natural population of Pinus taeda L. Heredity 107(2):105–114
Dhanapal AP, Ray JD, Singh SK, Hoyos-Villegas V, Smith JR, Purcell LC, King CA, Cregan PB, Song QJ, Fritschi FB (2015) Genome-wide association study (GWAS) of carbon isotope ratio (delta C-13) in diverse soybean [Glycine max (L.) Merr.] genotypes. Theor Appl Genet 128(1):73–91. https://doi.org/10.1007/s00122-014-2413-9
Easlon HM, Bloom AJ (2014) Easy leaf area: automated digital image analysis for rapid and accurate measurement of leaf area. Appl Plant Sci 2(7). https://doi.org/10.3732/apps.1400033
Easlon HM, Nemali KS, Richards JH, Hanson DT, Juenger TE, McKay JK (2013) The physiological basis for genetic variation in water use efficiency and carbon isotope composition in Arabidopsis thaliana. Photosynth Res 119(1):119–129. https://doi.org/10.1007/s11120-013-9891-5
Falconer DS, MacKay TFC (1996) Introduction to quantitative genetics. 4th Longman Essex, UK
Farquhar GD, Ehleringer JR, Hubick KT (1989) Carbon isotope discrimination and photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 40:503–537. https://doi.org/10.1146/Annurev.Arplant.40.1.503
Fisher RA (1918) The correlation between relatives on the supposition of Mendelian inheritance. Trans R Soc Edinb 52(02):399–433
Fulton AE, Buchner RP (2015) Drought tip: drought strategies for California walnut production.
Gelman A (2006) Prior distributions for variance parameters in hierarchical models (comment on an article by Browne and Draper). Bayesian Anal 1(3):515–533
Gelman A, Rubin DB (1992) Inference from iterative simulation using multiple sequences. Stat Sci 7(4):457–472
Goddard ME, Hayes BJ (2007) Genomic selection. J Anim Breed Genet 124(6):323–330
Gonzalez-Martinez SC, Huber D, Ersoz E, Davis JM, Neale DB (2008) Association genetics in Pinus taeda L. II. Carbon isotope discrimination. Heredity 101(1):19–26. https://doi.org/10.1038/hdy.2008.21
Guerra FP, Richards JH, Fiehn O, Famula R, Stanton BJ, Shuren R, Sykes R, Davis MF, Neale DB (2016) Analysis of the genetic variation in growth, ecophysiology, and chemical and metabolomic composition of wood of Populus trichocarpa provenances. Tree Genet Genomes 12(1):1–16. https://doi.org/10.1007/s11295-015-0965-8
Handley LL, Austin AT, Robinson D, Scrimgeour CM, Raven JA, Heaton THE, Schmidt S, Stewart GR (1999) The 15N natural abundance (δ15N) of ecosystem samples reflects measures of water availability. Aus J Plant Physiol 26(2):185–199. https://doi.org/10.1071/PP98146
Hikosaka K, Terashima I (1996) Nitrogen partitioning among photosynthetic component and its consequence in sun and shade plants. Funct Ecol 10(3):335–343
Hill WG, Goddard ME, Visscher PM (2008) Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet 4(2):e1000008
Huang D, Wu W, Abrams SR, Cutler AJ (2008) The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J Exp Bot 59(11):2991–3007. https://doi.org/10.1093/jxb/ern155
Impa SM, Nadaradjan S, Boominathan P, Shashidhar G, Bindumadhava H, Sheshshayee MS (2005) Carbon isotope discrimination accurately reflects variability in WUE measured at a whole plant level in rice. Crop Sci 45(6):2517–2522. https://doi.org/10.2135/cropsci2005.0119
Laity JH, Lee BM, Wright PE (2001) Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol 11(1):39–46. https://doi.org/10.1016/S0959-440x(00)00167-6
Lambers H, Chapin FS III, Pons TL (2008) Plant physiological ecology, 2nd edn. Springer-Verlag, New York
Lamesch P, Berardini TZ, Li DH, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, Karthikeyan AS, Lee CH, Nelson WD, Ploetz L, Singh S, Wensel A, Huala E (2012) The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40(D1):D1202–D1210. https://doi.org/10.1093/nar/gkr1090
Le Roux X, Sinoquet H, Vandame M (1999) Spatial distribution of leaf dry weight per area and leaf nitrogen concentration in relation to local radiation regime within an isolated tree crown. Tree Physiol 19(3):181–188
Marguerit E, Bouffier L, Chancerel E, Costa P, Lagane F, Guehl JM, Plomion C, Brendel O (2014) The genetics of water-use efficiency and its relation to growth in maritime pine. J Exp Bot 65(17):4757–4768
Marrano A, Martínez-García PJ, Bianco L, Sideli G, Di Pierro E, Leslie C, Stevens K, Crepeau M, Troggio M, Langley C, Neale DB (2018) A new genomic tool for walnut (Juglans regia L.): development and validation of the high-density Axiom™ J. regia 700K SNP genotyping array. Plant Biotechnol J. https://doi.org/10.1111/pbi.13034
Martínez-García PJ, Crepeau MW, Puiu D, Gonzalez-Ibeas D, Whalen J, Stevens KA, Paul R, Butterfield TS, Britton MT, Reagan RL, Chakraborty S, Walawage SL, Vasquez-Gross HA, Cardeno C, Famula RA, Pratt K, Kuruganti S, Aradhya MK, Leslie CA, Dandekar AM, Salzberg SL, Wegrzyn JL, Langley CH, Neale DB (2016) The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J 87(5):507–532. https://doi.org/10.1111/tpj.13207
Martínez-García PJ, Famula RA, Leslie C, McGranahan GH, Famula TR, Neale DB (2017) Predicting breeding values and genetic components using generalized linear mixed models for categorical and continuous traits in walnut (Juglans regia). Tree Genet Genomes 13(5):109. https://doi.org/10.1007/s11295-017-1187-z
McGranahan G, Leslie C (2012) Walnut. Fruit breeding 8:827–846. https://doi.org/10.1007/978-1-4419-0763-9_22
McKay J, Richards J, Mitchell-Olds T (2003) Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Mol Ecol 12(5):1137–1151
Niinements Ü (1999) Research review. Components of leaf dry mass per area – thickness and density – alter leaf photosynthetic capacity in reverse directions in woody plants. The New Phytologist 144(1):35–47
Powell JE, Visscher PM, Goddard ME (2010) Reconciling the analysis of IBD and IBS in complex trait studies. Nat Rev Genet 11(11):800–805. https://doi.org/10.1038/nrg2865
Prasolova NV, Xu ZH, Farquhar GD, Saffigna PG, Dieters MJ (2000) Variation in branchlet delta C-13 in relation to branchlet nitrogen concentration and growth in 8-year-old hoop pine families (Araucaria cunninghamii) in subtropical Australia. Tree Physiol 20(15):1049–1055
R Core Team (2017) R: a language and environment for statistical computing. R Foundation for statistical computing, Vienna URL https://www.R-project.org/
Ramos DE (1998) Walnut production manual. UC Davis. DANR. Communication Services Publications
Ramos DE, Brown LC, Uriu K, Marangoni B (1978) Water stress affects size and quality of walnuts. Calif Agric 32(10):5–6
Rebetzke G, Condon A, Farquhar G, Appels R, Richards R (2008) Quantitative trait loci for carbon isotope discrimination are repeatable across environments and wheat mapping populations. Theor Appl Genet 118(1):123–137. https://doi.org/10.1007/s00122-008-0882-4
Rosati A, Metcalf S, Buchner R, Fulton A, Lampinen B (2006) Tree water status and gas exchange in walnut under drought, high temperature and vapour pressure deficit. J Hortic Sci Biotechnol 81(3):415–420. https://doi.org/10.1080/14620316.2006.11512082
Seibt U, Rajabi A, Griffiths H, Berry JA (2008) Carbon isotopes and water use efficiency: sense and sensitivity. Oecologia 155(3):441–454. https://doi.org/10.1007/s00442-007-0932-7
Stevens KA, Woeste K, Chakraborty S, Crepeau MW, Leslie CA, Martínez-García PJ, Puiu D, Romero-Severson J, Coggeschall M, Dandekar AM, Kluepfel D, Neale DB, Salzberg SL, Langley CH (2018) Genomic variation among and within six Juglans species. G3: Genes Genom Genet:g3–g200030
Stich B, Mohring J, Piepho HP, Heckenberger M, Buckler ES, Melchinger AE (2008) Comparison of mixed-model approaches for association mapping. Genetics 178(3):1745–1754. https://doi.org/10.1534/genetics.107.079707
Stine S (1994) Extreme and persistent drought in California and Patagonia during medieval time. Nature 369(6481):546–549. https://doi.org/10.1038/369546a0
Thompson EA, Shaw RG (1990) Pedigree analysis for quantitative traits – variance-components without matrix-inversion. Biometrics 46(2):399–413. https://doi.org/10.2307/2531445
Tsai HY, Hamilton A, Tinch AE, Guy DR, Gharbi K, Stear MJ, Matika O, Bishop SC, Houston RD (2015) Genome wide association and genomic prediction for growth traits in juvenile farmed Atlantic salmon using a high density SNP array. BMC Genomics 16(1):969
Tulecke W, McGranahan G (1994) The walnut germplasm collection of the UC Davis. DANR, UC Davis
Turner SD (2014) qqman: An R package for visualizing GWAS results using QQ and Manhattan plots. BioRxiv, 005165
USDA NASS (2016) 2015 California walnut acreage report. National Agricultural Statistics Service 1–4
USDA-NASS (2017) Walnut/Raisin/Prune Report, State Summary- 2016 Crop Year. National Agricultural Statistics Service 1–4
Van Nocker S, Gardiner SE (2014) Breeding better cultivars, faster: applications of new technologies for the rapid deployment of superior horticultural tree crops. Hortic Res 1:14022. https://doi.org/10.1038/hortres.2014.22
Visscher PM, Hill WG, Wray NR (2008) Heritability in the genomics era – concepts and misconceptions. Nat Rev Genet 9(4):255–266. https://doi.org/10.1038/nrg2322
Wang D, Guo YH, Wu CG, Yang GD, Li YY, Zheng CC (2008) Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 9:44. https://doi.org/10.1186/1471-2164-9-44
Wimmer V, Albrecht T, Auinger HJ, Schon CC (2012) Synbreed: a framework for the analysis of genomic prediction data using R. Bioinformatics 28(15):2086–2087. https://doi.org/10.1093/bioinformatics/bts335
Acknowledgements
We would also like to thank Chuck Leslie and the Walnut Improvement Program for providing guidance with sampling and the Stable Isotope Facility at the University of California, Davis, for their laboratory work.
Data archiving statement
Raw phenotypic data used in trait analysis is available in the supplemental information. SNP data archived at Hardwood Genomics (https://www.hardwoodgenomics.org/Genome-assembly/2539069).
Funding
We would especially like to thank the California Walnut Board for supporting this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by S. C. González-Martínez
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Famula, R.A., Richards, J.H., Famula, T.R. et al. Association genetics of carbon isotope discrimination and leaf morphology in a breeding population of Juglans regia L.. Tree Genetics & Genomes 15, 6 (2019). https://doi.org/10.1007/s11295-018-1307-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-018-1307-4