Abstract
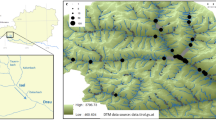
Riparian landscapes are dendritic in nature. However, much attention on genetic structure of riparian plants has been paid to linear models of connectivity while studies that investigate the influence of dendritic landscape are scarce. In this study, we used nuclear microsatellite markers to investigate genetic diversity, gene flow, and genetic structure of a streamside tree species (Euptelea pleiospermum) in a natural stream dendritic network in the Shennongjia Mountains, central China. We tested the following hypotheses: (1) genetic diversity is higher at confluence than that at headwater populations and (2) genetic structure within the stream dendritic network was determined by in-stream dispersal or out-of-stream dispersal. Contrary to our prediction, we found that both genetic diversity and effective population size are congruent at headwater and confluence populations. We found symmetrical gene flow in most (four out of six) headwater–confluence pairs and asymmetrically downstream gene flow in the other two headwater–confluence pairs. Analysis of molecular variance (AMOVA) detected significant differentiation at two scales (among streams within catchments, among populations within stream) and did not reveal significant structure among catchments. STRUCTURE analysis clustered individuals from different catchments into the same genetically homogeneous group. There was no significant isolation by distance (IBD) with Euclidean, stream, or overland distance. Our results suggest that E. pleiospermum populations within the stream dendritic network did not present a hierarchical genetic structure probably because of extensive out-of-stream dispersal.




Similar content being viewed by others
References
Baguette M, Blanchet S, Legrand D, Stevens VM, Turlure C (2013) Individual dispersal, landscape connectivity and ecological networks. Biol Rev 88:310–326
Beerli P, Felsenstein J (2001) Maximum likelihood estimation of a migration matrix and effective population size in n subpopulations by using a coalescent approach. Proc Natl Acad Sci U S A 98:4563–4568
Campbell Grant EH, Lowe WH, Fagan WF (2007) Living in the branches: population dynamics and ecological processes in dendritic networks. Ecol Let 10:165–175
Chaput-Bardy A, Fleurant C, Lemaire C, Secondi J (2009) Modelling the effect of in-stream and overland dispersal on gene flow in river networks. Ecol Model 220:3589–3598
Chaput-Bardy A, Lemaire C, Picard D, Secondi J (2008) In-stream and overland dispersal across a river network influences gene flow in a freshwater insect, Calopteryx splendens. Mol Ecol 17:3496–3505
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Crawford NG (2010) SMOGD: software for the measurement of genetic diversity. Mol Ecol Resour 10:556–557
Cushman SA, Landguth EL (2010) Scale dependent inference in landscape genetics. Landscape Ecol 25:967–979
Cushman SA, Max T, Meneses N, Evans LM, Ferrier S, Honchak B, Whitham TG, Allan GJ (2014) Landscape genetic connectivity in a riparian foundation tree is jointly driven by climatic gradients and river networks. Ecol Appl 24:1000–1014
Ehrich D (2009) Documentation for Structure-sum Version 2009. A series of R functions for summarizing the outputs of the program Structure ver. 2.2. http://uit.no/ansatte/organisasjon/ansatte/person?p_document_id=41186
Endress PK (1986) Floral structure, systematics and phylogeny of Trochodendrales. Ann Mo Bo Gard 73:297–324
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fagan WF (2002) Connectivity, fragmentation, and extinction risk in dendritic metapopulations. Ecology 83:3243–3249
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Fér T, Hroudová Z (2008) Detecting dispersal of Nuphar lutea in river corridors using microsatellite markers. Freshwater Biol 53:1409–1422
Fér T, Hroudová Z (2009) Genetic diversity and dispersal of Phragmites australis in a small river system. Aquat Bot 90:165–171
Fu LK, Jin JM (eds) (1992) China plant red data book: rare and endangered plants. Science Press, Beijing, pp 680–681
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices, version 2.9.3.2, updated from Goudet (1995). Available from http://www2.unil.ch/popgen/softwares/fstat.htm
Goudet J (2005) Hierfstat, a package for R to compute and test variance components and F-statistics. Mol Ecol Notes 5:184–186
Honnay O, Jacquemyn H, Nackaerts K, Breyne P, Van Looy K (2010) Patterns of population genetic diversity in riparian and aquatic plant species along rivers. J Biogeogr 37:1730–1739
Hopken MW, Douglas MR, Douglas ME (2013) Stream hierarchy defines riverscape genetics of a North American desert fish. Mol Ecol 22:956–971
Hu LJ, Uchiyama K, Shen HL, Ide Y (2010) Multiple-scaled spatial genetic structure of Fraxinus mandshurica over a riparian-mountain landscape in Northeast China. Conserv Genet 11:77–87
Hughes JM, Schmidt DJ, Finn DS (2009) Genes in streams: using DNA to understand the movement of freshwater fauna and their riverine habitat. Bioscience 59:573–583
Imbert E, Lefèvre F (2003) Dispersal and gene flow of Populus nigra (Salicaceae) along a dynamic river system. J Ecol 91:447–456
Jiang MX, Deng HB, Cai QH (2002) Distribution pattern of rare plants along riparian zone and its implication for conservation in Shennongjia area. Chinese J Appl Ecol 13:1373–1376
Johansson ME, Nilsson C, Nilsson E (1996) Do rivers function as corridors for plant dispersal? J Veg Sci 7:593–598
Jost L (2008) G ST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Kalinowski ST, Meeuwig MH, Narum SR, Taper ML (2008) Stream tree: a statistical method for mapping genetic differences between populations of freshwater organisms to the sections of streams that connect them. Can J Fish Aquat Sci 65:2752–2760
Kikuchi S, Suzuki W, Sashimura N (2009) Gene flow in an endangered willow Salix hukaoana (Salicaceae) in natural and fragmented riparian landscapes. Conserv Genet 12:79–89
Langella O (1999) Populations 1.2.32. Available via http://bioinformatics.org/~tryphon/populations/
Liu Y, Wang Y, Huang H (2006) High interpopulation genetic differentiation and unidirectional linear migration patterns in Myricaria laxiflora (Tamaricaceae), an endemic riparian plant in the Three Gorges Valley of the Yangtze River. Am J Bot 93:206–215
López-Pujol J, Zhang FM, Ge S (2006) Plant biodiversity in China: richly varied, endangered, and in need of conservation. Biodivers Conserv 15:3983–4026
López-Pujol J, Zhang FM, Sun HQ, Ying TS, Ge S (2011) Centres of plant endemism in China: places for survival or for speciation? J Biogeogr 38:1267–1280
Luikart G, Cornuet JM (1998) Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv Biol 12:228–237
Meffe GK, Vrijenhoek RC (1988) Conservation genetics in the management of desert dishes. Conserv Biol 2:157–169
Mitsui Y, Isagi Y, Setoguchi H (2010) Multiple spatial scale patterns of genetic diversity in riparian populations of Ainsliaea faurieana (Asteraceae) on Yakushima Island, Japan. Am J Bot 97:101–110
Morrissey MB, de Kerckhove DT (2009) The maintenance of genetic variation due to asymmetric gene flow in dendritic metapopulations. Am Nat 174:875–889
Mullen LB, Woods HA, Schwartz MK, Sepulveda AJ, Lowe WH (2010) Scale-dependent genetic structure of the Idaho giant salamander (Dicamptodon atterimus) in stream networks. Mol Ecol 19:898–909
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenic tree from molecular data. J Mol Evol 19:153–170
Nilsson C, Brown RL, Jansson R, Merritt DM (2010) The role of hydrochory in structuring riparian and wetland vegetation. Biol Rev 85:837–858
Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Galdstone J, Goyal R, Hakobsson M, Kim S, Morozov Y, Padhukasahasram B, Plagnol V, Rosenberg NA, Shah C, Wall JD, Wang J, Zhao K, Kalbfleisch T, Schulz V, Kreitman M, Bergelson J (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3, e196
Ohsawa T, Saito Y, Sawada H, Ide Y (2008) Impact of altitude and topography on the genetic diversity of Quercus serrata populations in the Chichibu Mountains, central Japan. Flora 203:187–196
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Pan KY, Lu AM, Wen J (1991) Chromosome number and development of gametophytes in Euptelea pleiospermum (Eupteleaceae). Acta Phytotaxon Sin 29:439–444
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioformatics 28:2537–2539
Peterson EE, Ver Hoef JM, Isaak DJ, Falke JA, Fortin MJ, Jordan CE, McNyset K, Monestiez P, Ruesch AS, Sengupta A, Som N, Steel EA, Theobald DM, Torgersen CE, Wenger SJ (2013) Modelling dendritic ecological networks in space: an integrated network perspective. Ecol Let 16:707–719
Phillipsen IC, Lytle DA (2013) Aquatic insects in a sea of desert: population genetic structure is shaped by limited dispersal in a naturally fragmented landscape. Ecography 36:731–743
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Pollux BJA, Luteijn A, van Groenendael JM, Ouborg NJ (2009) Gene flow and genetic structure of the aquatic macrophyte Sparganium emersum in a linear unidirectional river. Freshwater Biol 54:64–76
Prentis PJ, Mather PB (2008) Fine-scale patterns of genetic variation indicate non-equilibrium gene frequency divergence in the stream lily, Helmholtzia glaberrima. Freshwater Biol 53:973–980
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falush D (2009) Documentation for STRUCTURE software: version 2.3. Available from http://pritch.bsd.uchicago.edu/structure.html
Rice WR (1989) Analyzing tables of statistical tests. Evolution 3:23–225
Ritland K (1989) Genetic differentiation, diversity, and inbreeding in the mountain monkeyflower (Mimulus caespitosus) of the Washington Cascades. Can J Bot 67:2017–2024
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Sato T, Isagi Y, Sakio H, Osumi K, Goto S (2006) Effect of gene flow on spatial genetic structure in the riparian canopy tree Cercidiphyllum japonicum revealed by microsatellite analysis. Heredity 96:79–84
Tallmon DA, Koyuk A, Luikart G, Beaumont MA (2008) ONeSAMP: a program to estimate effective population size using approximate Bayesian computation. Mol Ecol Resour 8:299–301
Tero N, Aspi J, Siikamäki P, Jäkäläniemi A, Tuomi J (2003) Genetic structure and gene flow in a metapopulation of an endangered plant species, Silene tatarica. Mol Ecol 12:2073–2085
Tsuda Y, Sawada H, Ohsawa T, Nakao K, Nishikawa H, Ide Y (2010) Landscape genetic structure of Betula maximowicziana in the Chichibu mountain range, central Japan. Tree Genet Genomes 6:377–387
Waples RS (2006) A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv Genet 7:167–184
Waples RS, Do C (2008) LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8:753–756
Wei X, Jiang M (2012) Limited genetic impacts of habitat fragmentation in an “old rare” relict tree, Euptelea pleiospermum (Eupteleaceae). Plant Ecol 213:909–917
Wei X, Meng H, Jiang M (2013) Landscape genetic structure of a streamside tree species Euptelea pleiospermum (Eupteleaceae): contrasting roles of river valley and mountain ridge. PLoS ONE 8(6), e66928
Wei XZ, Huang HD, Jiang MX, Yang JY (2008) Quantitative characteristics and spatial distribution patterns of Euptelea pleiospermum populations in riparian zones of the Shennongjia area, central China. Chinese J Plant Ecol 32:825–837
Wei XZ, Jiang MX, Huang HD, Yang JY, Yu J (2010) Relationships between environment and mountain riparian plant communities associated with two rare tertiary-relict tree species, Euptelea pleiospermum (Eupteleaceae) and Cercidiphyllum japonicum (Cercidiphyllaceae). Flora 205:841–852
Weir B, Cockerham C (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wright S (1943) Isolation by distance. Genetics 28:114–138
Ying TS (2001) Species diversity and distribution pattern of seed plants in China. Biodivers Sci 9:393–398
Zhang J, Yao X, Wei X, Chen L, Jiang M (2008) Development and characterization of 14 polymorphic microsatellite loci in the endangered tree Euptelea pleiospermum (Eupteleaceae). Mol Ecol Resour 8:314–316
Acknowledgments
We are grateful for the assistance provided by Jianqing Ding, Mingxun Ren, Juan Yan, Fei Xiao, and Shijun Li. We also thank Dr. Feng Liu for reading and commenting on earlier draft of this manuscript. We appreciate two anonymous reviewers for their valuable comments and suggestions. Financial support was provided by the National Natural Science Foundation of China (Grant nos. 31100344 and 31270562), Key Laboratory of Aquatic Botany and Watershed Ecology, CAS (Grant no. Y455432J02), and the Youth Innovation Promotion Association, CAS (2014314).
Data archiving statement
Our microsatellite raw data for 240 Euptelea pleiospermum genotypes will be submitted to TreeGenes database.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. N. Aitken
This article is part of the Topical Collection on Population structure
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix 1
(DOCX 17 kb)
Rights and permissions
About this article
Cite this article
Wei, X., Meng, H., Bao, D. et al. Gene flow and genetic structure of a mountain riparian tree species, Euptelea pleiospermum (Eupteleaceae): how important is the stream dendritic network?. Tree Genetics & Genomes 11, 64 (2015). https://doi.org/10.1007/s11295-015-0886-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0886-6