Abstract
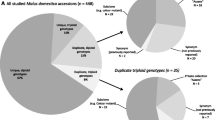
Potential interspecific hybrids are usually identified in natural populations by their proximity to interbreeding species or their intermediate phenotypes; hybridization can then be confirmed by comparing the genetic makeup of putative hybrids to pure species. In contrast, detecting interspecific hybridization and misclassifications in ex situ collections can be difficult because fine-scale geographic locations and species-specific phenotypic data are generally unavailable. Thus, there is little a priori information available to suggest which individuals might be hybrids. Instead, hybrids or misclassified individuals must be identified based on molecular data via population assignment and admixture detection programs. We have applied a variety of population assignment and admixture detection programs to over 400 samples of four closely related Malus species held in the US Department of Agriculture–Agricultural Research Service National Plant Germplasm System that were genotyped at 19 simple sequence repeat loci. Our findings indicate that over 10 % of the samples of the wild species Malus sieversii and Malus orientalis and nearly 20 % of the samples of the wild species Malus sylvestris may be admixed or misclassified. The percentage of admixed or misclassified samples of the domesticated species, Malus × domestica, was much lower, at <5 %. These findings provide an illustration of how to detect hybridization and misclassification in ex situ collections using molecular data and, ultimately, should help to maximize the utility of the collections. In addition, the presence of wild-collected samples that show admixture with domesticated apple suggests that gene flow may be occurring from the crop into natural populations of the wild species.


Similar content being viewed by others
References
Anderson EC, Thompson EA (2002) A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160:1217–1229
Besnard G, Henry P, Wille L, Cooke D, Chapuis E (2007) On the origin of the invasive olives (Olea europaea L., Oleaceae). Heredity 99:608–619
Breton C, Tersac M, Bervillé A (2006) Genetic diversity and gene flow between the wild olive (oleaster, Olea europaea L.) and the olive: several Plio-Pleistocene refuge zones in the mediterranean basin suggested by simple sequence repeats analysis. J Biogeogr 33:1916–1928. doi:10.1111/j.1365-2699.2006.01544.x
Buerkle CA (2005) Maximum-likelihood estimation of a hybrid index based on molecular markers. Mol Ecol Notes 5:684–687
Carney SE, Gardner KA, Rieseberg LH (2000) Evolutionary changes over the fifty-year history of a hybrid population of sunflowers (Helianthus). Evolution 54:462–474
Corander J, Marttinen P (2006) Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol 15:2833–2843. doi:10.1111/j.1365-294X.2006.02994.x
Corander J, Marttinen P, Siren J, Tang J (2008) Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9:539
De Carvalho D, Ingvarsson PK, Joseph J, Suter L, Sedivy C, Macaya-Sanz D, Cottrell J, Heinze B, Schanzer I, Lexer C (2010) Admixture facilitates adaptation from standing variation in the European aspen (Populus tremula L.), a widespread forest tree. Mol Ecol 19:1638–1650. doi:10.1111/j.1365-294X.2010.04595.x
Di Vecchi-Staraz M, Laucou V, Bruno G, Lacombe T, Gerber S, Bourse T, Boselli M, This P (2009) Low level of pollen-mediated gene flow from cultivated to wild grapevine: consequences for the evolution of the endangered subspecies Vitis vinifera L. subsp. silvestris. J Hered 100:66–75. doi:10.1093/jhered/esn084
Du FK, Petit RJ, Liu JQ (2009) More introgression with less gene flow: chloroplast vs. mitochondrial DNA in the Picea asperata complex in China, and comparison with other conifers. Mol Ecol 18:1396–1407. doi:10.1111/j.1365-294X.2009.04107.x
Duchesne P, Turgeon J (2009) FLOCK: a method for quick mapping of admixture without source samples. Mol Ecol Resour 9:1333–1344. doi:10.1111/j.1755-0998.2009.02571.x
Earl DA (2011) Structure harvester v0.6. Available at http://taylor0.biology.ucla.edu/struct_harvest/
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Field DL, Ayre DJ, Whelan RJ, Young AG (2011) Patterns of hybridization and asymmetrical gene flow in hybrid zones of the rare Eucalyptus aggregata and common E. rubida. Heredity 106:841–853. doi:10.1038/hdy.2010.127
Glaubitz JC (2004) CONVERT: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes 4:309–310
Guilford P, Prakash S, Zhu JM, Rikkerink E, Gardiner S, Bassett H, Forster R (1997) Microsatellites in Malus × domestica (apple): abundance, polymorphism and cultivar identification. Theor Appl Genet 94:249–254. doi:10.1007/s001220050407
Hokanson SC, Szewc-McFadden AK, Lamboy WF, McFerson JR (1998) Microsatellite (SSR) markers reveal genetic identities, genetic diversity and relationships in a Malus × domestica borkh. core subset collection. Theor Appl Genet 97:671–683. doi:10.1007/s001220050943
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. doi:10.1093/bioinformatics/btm233
Korban SS, Skirvin RM (1984) Nomenclature of the cultivated apple. Hortscience 19:177–180
Kovach MJ, McCouch SR (2008) Leveraging natural diversity: back through the bottleneck. Curr Opin Plant Biol 11:193–200
Langella O, Chikhi L, Beaumont MA (2001) LEA (likelihood-based estimation of admixture): a program to estimate simultaneously admixture and time since the admixture event. Mol Ecol Notes 1:357–358. doi:10.1046/j.1471-8278.2001.00099.x
Lepais O, Petit RJ, Guichoux E, Lavabre JE, Alberto F, Kremer A, Gerber S (2009) Species relative abundance and direction of introgression in oaks. Mol Ecol 18:2228–2242. doi:10.1111/j.1365-294X.2009.04137.x
Lexer C, Joseph JA, van Loo M, Barbara T, Heinze B, Bartha D, Castiglione S, Fay MF, Buerkle CA (2010) Genomic admixture analysis in European Populus spp. reveals unexpected patterns of reproductive isolation and mating. Genetics 186:699–712. doi:10.1534/genetics.110.118828
Liebhard R, Gianfranceschi L, Koller B, Ryder CD, Tarchini R, Van De Weg E, Gessler C (2002) Development and characterisation of 140 new microsatellites in apple (Malus × domestica Borkh.). Molecular Breeding 10:217–241. doi:10.1023/a:1020525906332
Luby J, Forsline P, Aldwinckle H, Bus V, Geibel M (2001) Silk road apples—collection, evaluation, and utilization of Malus sieversii from central Asia. HortScience 36:225–231
Miller AJ, Gross BL (2011) Forest to field: perennial fruit crop domestication. Am J Bot 98:1389–1414. doi:10.3732/ajb.1000522
Palmé AE, Vendramin GG (2002) Chloroplast DNA variation, postglacial recolonization and hybridization in hazel, Corylus avellana. Mol Ecol 11:1769–1779. doi:10.1046/j.1365-294X.2002.01581.x
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. Available at http://darwin.cirad.fr/darwin
Perrier X, Flori A, Bonnot F (2003) Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC (eds) Genetic diversity of cultivated tropical plants. Science Publishers, Montpellier, Enfield, pp 43–76
Petit RJ, Bodénès C, Ducousso A, Roussel G, Kremer A (2004) Hybridization as a mechanism of invasion in oaks. New Phytol 161:151–164. doi:10.1046/j.1469-8137.2003.00944.x
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rambaut A (2006) FigTree v1.3.1. 1.3.1 edn. Available at http://tree.bio.ed.ac.uk/
Remington DL, Robichaux RH (2007) Influences of gene flow on adaptive speciation in the Dubautia arborea–D. ciliolata complex. Mol Ecol 16:4014–4027. doi:10.1111/j.1365-294X.2007.03447.x
Richards C, Volk G, Reilley A, Henk A, Lockwood D, Reeves P, Forsline P (2009a) Genetic diversity and population structure in Malus sieversii, a wild progenitor species of domesticated apple. Tree Genet Genomes 5:339–347. doi:10.1007/s11295-008-0190-9
Richards CM, Volk GM, Reeves PA, Reilley AA, Henk AD, Forsline PL, Aldwinckle HS (2009b) Selection of stratified core sets representing wild apple (Malus sieversii). J Am Soc Hortic Sci 134:228–235
Rieseberg LH, Archer MA, Wayne RK (1999) Transgressive segregation, adaptation and speciation. Heredity 83:363–372
Robinson JP, Harris SA, Juniper BE (2001) Taxonomy of the genus Malus Mill. (Rosaceae) with emphasis on the cultivated apple, Malus domestica Borkh. Plant Syst Evol 226:35–58. doi:10.1007/s006060170072
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138. doi:10.1046/j.1471-8286.2003.00566.x
Routson KJ, Reilley AA, Henk AD, Volk GM (2009) Identification of historic apple trees in the Southwestern United States and implications for conservation. HortScience 44:589–594
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–426
Sanz N, Araguas R, Fernández R, Vera M, García-Marín J-L (2009) Efficiency of markers and methods for detecting hybrids and introgression in stocked populations. Conserv Genet 10:225–236. doi:10.1007/s10592-008-9550-0
Szymura JM, Barton NH (1991) The genetic structure of the hybrid zone between the fire-bellied toads Bombina bombina and B. variegata: comparisons between transects and between loci. Evolution 45:237–261
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066. doi:10.1126/science.277.5329.1063
Vähä J-P, Primmer CR (2006) Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol Ecol 15:63–72. doi:10.1111/j.1365-294X.2005.02773.x
Velasco R, Zharkikh A, Affourtit J (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–839, http://www.nature.com/ng/journal/vaop/ncurrent/abs/ng.654.html-supplementary-information
Volk GM, Richards CM, Reilley AA, Henk AD, Forsline PL, Aldwinckle HS (2005) Ex situ conservation of vegetatively propagated species: development of a seed-based core collection for Malus sieversii. J Am Soc Hortic Sci 130:203–210
Volk GM, Richards CM, Henk AD, Reilley AA, Reeves PA, Forsline PL, Aldwinckle HS (2009) Capturing the diversity of wild Malus orientalis from Georgia, Armenia, Russia, and Turkey. J Am Soc Hortic Sci 134:453–459
Wang H, McArthur ED, Sanderson SC, Graham JH, Freeman DC (1997) Narrow hybrid zone between two subspecies of big sagebrush (Artemisia tridentata: Asteraceae). IV. Reciprocal transplant experiments. Evolution 51:95–102
Yamamoto T, Kimura T, Shoda M, Ban Y, Hayashi T, Matsuta N (2002) Development of microsatellite markers in the Japanese pear (Pyrus pyrifolia Nakai). Mol Ecol Notes 2:14–16. doi:10.1046/j.1471-8286.2002.00128.x
Acknowledgments
We thank Ann A. Reilley, Celeste Falcon, and Amy K. Szewc-McFadden for their assistance in the data collection for this project. We thank Jared L. Strasburg and Christina T. Walters for their comments on a previous draft of this manuscript. This project was partially supported by the National Research Initiative of the United States Department of Agriculture Cooperative State Research, Education, and Extension Service, grant number #2005-00751.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Dirlewanger
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online resource 1
Species assignments and admixture analyses from all separate analyses. See color coding for key. The two columns at the end of the table (Admixed Overall; Admixed with Overall) correspond to the results in Table 2 (PDF 216 kb)
Online resource 2
Primer names, sequences, observed size range and number of alleles, approximate annealing temperature, repeat type, map location, and reference for each SSR (PDF 174 kb)
Online resource 3
Graphical representations of results from STRUCTURE, BAPS, FLOCK, and DARwin conducted on the full dataset and the dataset excluding known families. For STRUCTURE, BAPS, and FLOCK, species groups, analysis method and levels of K indicated are below each plot. Each bar (not delineated) represents a sample, and the colors represent the estimated membership in the genetic clusters identified by the programs. Species groups are delineated with black lines. There are three graphical representations of the STRUCTURE results for K = 4 without families; one shows the average of all 10 runs, one shows the average of six runs that converged on one solution and one shows the average of four runs that converged on alternate solution. For DARwin, each sample is represented as a terminal node in the neighbor-joining tree, and the samples are color-coded according to their original species designations in GRIN; clades of species are labeled with text. *K was set equal to four for the BAPS analyses, so the level of K was not estimated by the program (PDF 598 kb)
Online resource 4
Allele frequencies for each species and overall allele frequencies for all 19 SSR loci, calculated after the removal of all misclassified or admixed samples from the dataset. See color coding for key. Note that the total number of samples will differ for each locus due to missing data (XLS 274 kb)
Online resource 5
All available photos of admixed or misclassified samples, as well as a representative sample of each species, labeled with PI number, cultivar name (if applicable), currently assigned species status, and species they are admixed with in parentheses (dom = M. × domestica, ori = M. orientalis, sie = M. sieversii, syl = M. sylvestris). Images are resized to roughly the same scale (1 cm grid is shown with dots in the background of each photo) (PDF 3986 kb)
Rights and permissions
About this article
Cite this article
Gross, B.L., Henk, A.D., Forsline, P.L. et al. Identification of interspecific hybrids among domesticated apple and its wild relatives. Tree Genetics & Genomes 8, 1223–1235 (2012). https://doi.org/10.1007/s11295-012-0509-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-012-0509-4