Abstract
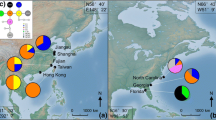
Studies of genetic variation can elucidate the structure of present and past populations as well as the genetic basis of the phenotypic variability of species. Taxodium distichum is a coniferous tree dominant in lowland river flood plains and swamps of the southeastern USA which exhibits morphological variability and adaption to stressful habitats. This study provides a survey of the Mississippi River Alluvial Valley (MAV) and Florida to elucidate their population structure and the extent of genetic differentiation between the two regions and sympatric varieties, including bald cypress (var. distichum) and pond cypress (var. imbricatum). We determined the genotypes of 12 simple sequence repeat loci totaling 444 adult individuals from 18 natural populations. Bayesian clustering analysis revealed high levels of differentiation between the MAV and the Florida regions. Within the MAV region, there was a significant correlation between genetic and geographical distances. In addition, we found that there was almost no genetic differentiation between the varieties. Most genetic variation was found within individuals (76.73 %), 1.67 % among individuals within population, 15.36 % among populations within the regions, and 9.23 % between regions within the variety. Our results suggest that (1) the populations of the MAV and the Florida regions are divided into two major genetic groups, which might originate from different glacial refugia, and (2) the patterns of genetic differentiation and phenotypic differentiation were not parallel in this species.





Similar content being viewed by others
References
Al-Rabab’ah MA, Williams CG (2002) Population dynamics of Pinus taeda L. based on nuclear microsatellites. Forest Ecol Manage 163:263–271
Armour JA, Neumann R, Gobert S, Jeffreys AJ (1994) Isolation of human simple repeat loci by hybridization selection. Hum Mol Genet 3(4):599–605
Avise JC (1989) Gene trees and organismal histories: a phylogenetic approach to population biology. Evolution 43(6):1192–1208
Brown S (1981) A comparison of the structure, primary productivity, and transpiration of cypress ecosystems in Florida. Ecol Monogr 51:403–427
Callen DF, Thompson AD, Shen Y, Philips HA, Richards RI, Mulley JC, Sutherland GR (1993) Incidence and origin of “null” alleles in the (AC) n microsatellite markers. Am J Hum Genet 52:922–927
Delcourt PA, Delcourt HR (1981a) Vegetation maps for eastern North America: 20 000 yr. BP to the present. In: Romans RC (ed) Geobotany. Plenum, New York, pp 123–165
Delcourt PA, Delcourt HR (1981b) Vegetation maps for eastern North America: 40,000 yr b.p. to the present. In: Romans RC (ed) Geobotany. Plenum, New York, p 263
Duminil J, Hardy OJ, Petit RJ (2009) Plant traits correlated with generation time directly affect inbreeding depression and mating system and indirectly genetic structure. BMC Evol Biol 9:177
Duran C, Appleby N, Edwards D, Batley J (2009) Molecular genetic markers: discovery, applications, data storage and visualisation. Curr Bioinform 4:16–27
Eckert CG, Samis KE, Lougheed SC (2008) Genetic variation across species’ geographical ranges: the central–marginal hypothesis and beyond. Mol Ecol 17(5):1170–1188
Elsik CG, Williams CG (2001) Low-copy microsatellite recovery from a conifer genome. Theor Appl Genet 103(8):1189–1195
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3):564–567
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131(2):479–491
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Farjon A (2005) A monograph of Cupressaceae and Sciadopitys. Kew Publishing, Richmond
Fleischer RC, Loew S (1995) Construction and screening of microsatellite-enriched genomic libraries. In: Ferraris J, Palumbi S (eds) Molecular zoology: advances, strategies and protocols. Wiley, New York, pp 459–468
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86(6):485–486
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (ver. 2.9.3). Computer program and documentation distributed by the author. http://www2.unil.ch/popgen/softwares/fstat/htm. Accessed Apr 2011
Goudet J (2005) Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186
Hampe A, Petit RJ (2005) Conserving biodiversity under climate change: the rear edge matters. Ecol Lett 8(5):461–467
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New Forests 6:95–124
Hancock JM (1999) Microsatellites and other simple sequences: genomic context and mutational mechanisms. In: Goldstein DB, Schlötterer C (eds) Microsatellites: evolution and applications. Oxford University Press, Oxford, pp 1–9
Hewitt GM (1999) Post-glacial re-colonization of European biota. Biol J Linn Soc 68(1–2):87–112
Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philos Trans Royal Soc London Ser B: Biol Sci 359(1442):183–195
Hey J (2010) Isolation with migration models for more than two populations. Mol Biol Evol 27(4):905–920
Jaramillo-Correa JP, Beaulieu J, Khasa DP, Bousquet J (2009) Inferring the past from the present phylogeographic structure of North American forest trees: seeing the forest for the genes. Can J For Res 39(2):286–307
Jaramillo-Correa JP, Verdú M, González-Martíne SC (2010) The contribution of recombination to heterozygosity differs among plant evolutionary lineages and life-forms. BMC Evol Biol 10:22
Kado T, Ushio Y, Yoshimaru H, Tsumura Y, Tachida H (2006) Contrasting patterns of DNA variation in natural populations of two related conifers, Cryptomeria japonica and Taxodium distichum (Cupressaceae sensu lato). Genes Genet Syst 81(2):103–113
Kusumi J, Zidong L, Kado T, Tsumura Y, Middleton BA, Tachida H (2010) Multilocus patterns of nucleotide polymorphism and demographic change in Taxodium distichum (Cupressaceae) in the lower Mississippi River Alluvial Valley. Am J Bot 97(11):1848–1857
Langella O (2002) Population 1.2.30. Computer program and documentation distributed by the author. http://bioinformatics.org/~tryphon/populations/. Accessed Nov 2009
Lickey EB, Walker GL (2002) Population genetic structure of baldcypress (Taxodium distichum [L.] Rich. var. distichum) and pondcypress (T. distichum var. imbricarium [Nuttall] Croom). Southeast Nat 1(2):131–148
Mantel NA (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Middleton BA, McKee KL (2004) Use of a latitudinal gradient in bald cypress (Taxodium distichum) production to examine physiological controls of biotic boundaries and potential responses to environmental change. Glob Ecol Biogeogr 13(3):247–258
Moriguchi Y, Ueno S, Ujino-Ihara T, Futamura N, Matsumoto A, Shinohara K, Tsumura Y (2009) Characterization of EST-SSRs from Cryptomeria japonica. Conserv Genet Resour 1(1):373–376
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19:153–170
Neufeld HS (1986) Ecophysiological implications of tree architecture for two cypress taxa, Taxodium distichum (L.) Rich. and T. ascendens Brongn. Bull Torrey Bot Club 113:118–124
Nielsen R, Beaumont MA (2009) Statistical inferences in phylogeography. Mol Ecol 18(6):1034–1047
Peakall R, Smouse PE (2006) GENALEX6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Petit RJ, Aguinagalde I, de Beaulieu JL, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, Mohanty A, Muller-Starck G, Demesure-Musch B, Palme A, Martin JP, Rendell S, Vendramin GG (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300(5625):1563–1565
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Sagarin RD, Gaines SD (2002) The ‘abundant centre’ distribution: to what extent is it a biogeographical rule? Ecol Lett 5(1):137–147
Sagarin RD, Gaines SD, Gaylord B (2006) Moving beyond assumptions to understand abundance distributions across the ranges of species. Trends Ecol Evol 21(9):524–530
Schmidtling RC, Hipkins V (1998) Genetic diversity in long-leaf pine (Pinus palustris): influence of historical and prehistorical events. Can J Forest Res 28:1135–1145
Soltis DE, Morris AB, MacLachlan JS, Manos PS, Soltis PS (2006) Comparative phylogeography of unglaciated eastern North America. Mol Ecol 15:4261–4293
Strasburg JL, Sherman NA, Wright KM, Moyle LC, Willis JH, Rieseberg LH (2012) What can patterns of differentiation across plant genomes tell us about adaptation and speciation? Philos Trans Royal Soc B 367:364–373
Takahashi T, Tani N, Taira H, Tsumura Y (2005) Microsatellite markers reveal high allelic variation in natural populations of Cryptomeria japonica near refugial areas of the last glacial period. J Plant Res 118(2):83–90
Tsumura Y, Tomaru N, Suyama Y, Bacchus S (1999) Genetic diversity and differentiation of Taxodium in the south-eastern United States using cleaved amplified polymorphic sequences. Heredity 83:229–238
Watson FD (1986) The nomencalture of pondcypress and bald-cypress (Taxodiaceae). Taxon 34:506–509
Watts WA (1973) The vegetation record of a Mid-Wisconsin interstadial in northwest Georgia. Quanternary Res 3:257–268
Wilhite LP, Toliver JR (1990) Taxodium distichum (L.) Rich. baldcypress. In: Burns RM, Honkala BH (eds) Silvics of North America: 1. Conifers. U.S. Department of Agriculture, Forest Service, Washington, DC
Wright S (1943) Isolation by distance. Genetics 28(2):114–138
Wright S (1951) The general structure of populations. Ann Eugen 15:323–354
Yang RC (1998) Estimating hierarchical F-statistics. Evolution 52(4):950–956
Acknowledgments
The authors thank Y. Moriguchi and A. Sato for their help with the development of SSR markers and DNA extraction. We also thank S. Travis, J. Grace, and an anonymous reviewer for helpful suggestions on earlier drafts of the manuscript. This study was partially supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (no. 22370083) and by Program for promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN). Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the US Government.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Sederoff
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Appendix S1
Characterization of twelve T. distichum SSR marker loci, including accession numbers, primer sequences, motif, annealing temperatures (T a) (DOC 60.5 kb)
Appendix S3
Pairwise F st values of 18 populations. All pair showed a significant genetic differentiation (P < 0.01) (DOC 61 kb)
Appendix S4
Analysis of molecular variance (AMOVA) of populations at the MAV region (bald cypress only) (A) and at Florida region (B) (DOC 43.5 kb)
Rights and permissions
About this article
Cite this article
Tanaka, A., Ohtani, M., Suyama, Y. et al. Population genetic structure of a widespread coniferous tree, Taxodium distichum [L.] Rich. (Cupressaceae), in the Mississippi River Alluvial Valley and Florida. Tree Genetics & Genomes 8, 1135–1147 (2012). https://doi.org/10.1007/s11295-012-0501-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-012-0501-z