Abstract
Flowering time is a well-studied subject in ecology, evolution and molecular biology. Long-term phenological studies have shown relationships between flowering time and environmental and endogenous factors in many species. In contrast, molecular studies using model plants have revealed a complex regulatory network of flowering. We propose that flowering would be a model trait for the integrated study of ecology, evolution and molecular biology. We introduce briefly the flowering regulatory pathways of Arabidopsis thaliana, followed by molecular techniques such as transgenic manipulation, quantitative real-time PCR and detection of differentially expressed genes that could facilitate the study of ‘nonmodel’ species of ecological interest but with little available genome information. Application of the flowering gene network to wild species will be illustrated by two examples: modeling and prediction of the expression of flowering genes in Arabidopsis halleri, and the latitudinal cline of bud set and cessation in Populus. Finally, we discuss the challenges in integrating knowledge of the regulatory network on flowering into ecologically unique flowering phenomena such as synchronous intermittent mass flowering—the topic of this special issue.



Similar content being viewed by others
References
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309:1052–1056
Aikawa S, Kobayashi MJ, Satake A, Shimizu KK, Kudoh H (2010) Robust control of the seasonal expression of the Arabidopsis FLC gene in a fluctuating environment. Proc Natl Acad Sci USA 22:11632–11637
Aizen MA (2003) Influences of animal pollination and seed dispersal on winter flowering in a temperate mistletoe. Ecology 84:2613–2627
Amasino R (2010) Seasonal and developmental timing of flowering. Plant J 61:1001–1013
Ashton PS, Givnish TJ, Appanah S (1988) Staggered flowering in the Dipterocarpaceae: new insights into floral induction and the evolution of mast fruiting in the aseasonal tropics. Am Nat 132:44–66
Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36:162–166
Balasubramanian S, Sureshkumar S, Lempe J, Weigel D (2006) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2:e106
Barakat A, DiLoreto DS, Zhang Y, Smith C, Baier K, Powell WA, Wheeler N, Sederoff R, Carlson JE (2009) Comparison of the transcriptomes of American chestnut (Castanea dentata) and Chinese chestnut (Castanea mollissima) in response to the chestnut blight infection. BMC Plant Biol 9:51
Bellin D, Ferrarini A, Chimento A, Kaiser O, Levenkova N, Bouffard P, Delledonne M (2009) Combining next-generation pyrosequencing with microarray for large scale expression analysis in non-model species. BMC Genomics 10:555
Birzele F, Schaub J, Rust W, Clemens C, Baum P, Kaufmann H, Weith A, Schulz TW, Hildebrandt T (2010) Into the unknown: expression profiling without genome sequence information in CHO by next generation sequencing. Nucleic Acids Res 38:3999–4010
Blázquez MA, Weigel D (2000) Integration of floral inductive signals in Arabidopsis. Nature 404:889–892
Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 19:1040–1043
Brearley FQ, Proctor J, Suriantata, Nagy L, Dalrymple G, Voysey BC (2007) Reproductive phenology over 10-year period in a lowland evergreen rain forest of central Borneo. J Ecol 95:828–839
Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25:169–193
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Nuller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Cerdán PD, Chory J (2003) Regulation of flowering time by light quality. Nature 19:881–885
Chan SK, Struhl G (1997) Sequence-specific RNA binding by Bicoid. Nature 388:634
Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 25:4985–4990
Chuck G, Cigan AM, Saeteurn K, Hake S (2007) The heterochronic maize mutant Congrass1 results from overexpression of a tandem microRNA. Nat Genet 39:544–549
Chuine I, Kramer K, Hanninen H (2003) Plant development models. In: Schwartz MD (ed) Phenology: an integrative environmental science. Kluwer, Dordrecht, pp 217–235
Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 18:1030–1033
Crone EE, Miller E, Sala A (2009) How do plants know when other plants are flowering? Resource depletion, pollen limitation and mast-seeding in a perennial wildflower. Ecol Lett 12:1119–1126
Dennis ES, Peacock WJ (2009) Vernalization in cereals. J Biol 8:57
Distelfeld A, Li C, Dubcovsky J (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12:178–184
Ekblom R, Balakrishnan CN, Burke T, Slate J (2010) Digital gene expression analysis of the zebra finch genome. BMC Genomics 11:219
Eriksson S, Böhlenius H, Moritz T, Nilsson O (2006) GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18:2172–2181
Fischer RA, Turner NC (1978) Plant productivity in the arid and semi-arid zones. Annu Rev Plant Physiol 29:277–317
Fornara F, de Montaigu A, Coupland G (2010) SnapShot: control of flowering in Arabidopsis. Cell 30:550
Frewen BE, Chen TH, Howe GT, Davis J, Rohde A, Boerjan W, Bradshaw HD Jr (2000) Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics 154:837–845
Greenup A, Peacock WJ, Dennis ES, Trevaskis B (2009) The molecular biology of seasonal flowering-responses in Arabidopsis and cereals. Ann Bot 103:1165–1172
Halliday KJ, Salter MG, Thingnaes E, Whitelam GC (2003) Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J 33:875–885
Hättasch C, Flachowsky H, Kapturska D, Hanke MV (2008) Isolation of flowering genes and seasonal changes in their transcript levels related to flower induction and initiation in apple (Malus domestica). Tree Physiol 28:1459–1466
He Y, Michaels SD, Amasino RM (2003) Regulation of flowering time by histone acetylation in Arabidopsis. Science 302:1751–1754
Helliwell CA, Wood CC, Robertson M, Peacock WJ, Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46:183–192
Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21:4327–4337
Higgins JA, Bailey PC, Laurie DA (2010) Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS One 19:e10065
Howe GT, Gardner G, Hackett WP, Furnier GR (1996) Phytochrome control of short-day-induced bud set in black cottonwood. Physiol Plant 97:95–103
Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11:550–558
Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286:1962–1965
Kawagoe T, Kudoh H (2010) Escape from floral herbivory by early flowering in Arabidopsis halleri subsp. gemmifera. Oecologia 164:713–720
Kelly D (1994) The evolutionary ecology of mast seeding. Trends Ecol Evol 9:465–470
Kelly D, Sork VL (2002) Mast seeding in perennial plants: why, how, where? Annu Rev Ecol Syst 33:427–447
Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286:1960–1962
Kobayashi Y, Weigel D (2007) Move on up, it’s time for change—mobile signals controlling photoperiod-dependent flowering. Genes Dev 1:2371–2384
Koenig WD, Knops JM (2000) Patterns of annual seed production by northern hemisphere trees: a global perspective. Am Nat 155:59–69
Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day condition. Plant Cell Physiol 43:1096–1105
Koornneef M, Hanhart CJ, Van Der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229:57–66
Kotoda N, Hayashi H, Suzuki M, Igarashi M, Hatsuyama Y, Kidou S, Igasaki T, Nishiguchi M, Yano K, Shimizu T, Takahashi S, Iwanami H, Moriya S, Abe K (2010) Molecular characterization of FLOWERING LOCUS T-like genes of apple (Malus × domestica Borkh.). Plant Cell Physiol 51:561–575
Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14:2366–2376
Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 15:397–402
Lee S, Kim J, Han JJ, Han MJ, An G (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO1/AGAMOUS-LIKE20 (SOC1/AGL20) ortholog in rice. Plant J 38:754–764
Li C, Dubcovsky J (2008) Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J 55:543–554
Lim MH, Kim J, Kim YS, Chung KS, Seo YH, Lee I, Hong CB, Kim HJ, Park CM (2004) A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16:731–740
Lloyd AM, Barnason AR, Rogers SG, Byrne MC, Fraley RT, Horsch RB (1986) Transformation of Arabidopsis thaliana with Agrobacterium tumefaciens. Science 24:464–466
Locascio A, Lucchin M, Varotto S (2009) Characterization of a MADS FLOWERING LOCUS C-LIKE (MFL) sequence in Cichorium intybus: a comparative study of CiMFL and AtFLC reveals homologies and divergences in gene function. New Phytol 182:630–643
Ma Y, Qi X, Du J, Song S, Feng D, Qi J, Zhu Z, Zhang X, Xiao H, Han Z, Hao X (2009) Identification of candidate genes for human pituitary development by EST analysis. BMC Genomics 10:109
Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 30:737–745
Masaki T, Oka T, Osumi K, Suzuki W (2008) Geographical variation in climatic cues for mast seeding of Fagus crenata. Popul Ecol 50:357–366
Matsumura H, Krüger DH, Kahl G, Terauchi R (2008) SuperSAGE: a modern platform for genome-wide quantitative transcript profiling. Curr Pharm Biotechnol 9:368–374
Meinke DW, Cherry JM, Dean C, Rounsley SD, Koornneef M (1998) Arabidopsis thaliana: a model plant for genome analysis. Science 23:679–682
Michaels SD (2009) Flowering time regulation produces much fruit. Curr Opin Plant Biol 12:75–80
Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11:949–956
Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13:935–942
Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol 137:149–156
Mitchell-Olds T, Schmitt J (2006) Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 22:947–952
Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35:613–623
Morinaga SI, Tsuji K, Sakai S (2003) Consequences of differences in flowering date on seed production in Heloniopsis orientalis (Liliaceae). Am J Bot 90:1153–1158
Mouhu K, Hytönen T, Folta K, Rantanen M, Paulin L, Auvinen P, Elomaa P (2009) Identification of flowering genes in strawberry, a perennial SD plant. BMC Plant Biol 28:122
Murai K, Miyamae M, Kato H, Takumi S, Ogihara Y (2003) WAP1, a wheat APETALA1 homolog, plays a central role in the phase transition from vegetative to reproductive growth. Plant Cell Physiol 44:1255–1265
Mutasa-Göttgens E, Hedden P (2009) Gibberellin as a factor in floral regulatory networks. J Exp Bot 60:1979–1989
Nishikawa F, Endo T, Shimada T, Fujii H, Shimizu T, Kobayashi Y, Araki T, Omura M (2010) Transcriptional changes in CiFT-introduced transgenic trifoliate orange (Poncirus trifoliate L. Raf.). Tree Physiol 30:431–439
Nishikawa F, Endo T, Shimada T, Fujii H, Shimizu T, Omura M (2009) Differences in seasonal expression of flowering genes between deciduous trifoliate orange and evergreen Satsuma mandarin. Tree Physiol 29:921–926
Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16:2601–2613
Nordborg M, Bergelson J (1999) The effect of seed and rosette cold treatment on germination and flowering time in some Arabidopsis thaliana (Brassicaceae) ecotypes. Am J Bot 86:470
Numata S, Yasuda M, Okuda T, Kachi N, Nur Supardi MN (2003) Temporal and spatial patterns of mass flowerings on the Malay peninsula. Am J Bot 90:1025–1031
O’Neil P (1999) Selection on flowering time: an adaptive fitness surface for nonexistent character combinations. Ecology 80:806–820
Opler PA, Frankie GW, Baker HG (1976) Rainfall as a factor in the release, timing, and synchronization of anthesis by tropical trees and shrubs. J Biogeogr 3:231–236
Ow DW, Wood KV, DeLuca M, De Wet JR, Helinski DR, Howell SH (1986) Transient and stable expression of the firefly luciferase gene in plant cells and transgenic plants. Science 14:856–859
Pauley SS, Perry TO (1954) Ecotypic variation in the photoperiodic response in Populus. J Arnold Arbor Harv Univ 35:167
Peña L, Martin-Trillo M, Juárez J, Pina JA, Navarro L, Martinez-Zapater JM (2001) Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat Biotechnol 19:263–267
Pirooznia M, Pozhitkov A, Perkins EJ, Deng Y, Brouwer M (2010) Generation, analysis and functional annotation of expressed sequence tags from the sheepshead minnow (Cyprinodon variegates). BMC Genomics 11(Suppl 2):4
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80:847–857
Rao NN, Prasad K, Kumar PR, Vijayraghavan U (2008) Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc Natl Acad Sci USA 4:3646–3651
Rees M, Kelly D, Bjørnstad O (2002) Snow tussocks, chaos, and the evolution of mast seeding. Am Nat 160:44–59
Reeves PA, He Y, Schmitz RJ, Amasino RM, Panella LW, Richards CM (2007) Evolutionary conservation of the FLOWERING LOCUS C-mediated vernalization response: evidence from the sugar beet (Beta vulgaris). Genetics 176:295–307
Reeves PH, Coupland G (2001) Analysis of flowering time control in Arabidopsis by comparison of double and triple mutants. Plant Physiol 126:1085–1091
Rodriguez MC, Edsgard D, Hussain SS, Alquezar D, Rasmussen M, Gilbert T, Nielsen BH, Bartels D, Mundy J (2010) Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. Plant J 63:212–228
Sakai S, Momose K, Yumoto T, Nagamitsu T, Nagamasu H, Hamid AA, Nakashizuka T (1999) Plant reproductive phenology over four years including an episode of general flowering in a lowland dipterocarp forest, Sarawak, Malaysia. Am J Bot 86:1414–1436
Sakai S, Harrison RD, Momose K, Kuraji K, Nagamasu H, Yasunari T, Chong L, Nakashizuka T (2006) Irregular droughts trigger mass flowering in aseasonal tropical forests in Asia. Am J Bot 93:1134–1139
Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288:1613–1616
Schmid J, Muller-Hagen D, Bekel T, Funk L, Stahl U, Sieber V, Meyer V (2010) Transcriptome sequencing and comparative transcriptome analysis of the scleroglucan producer Sclerotium rolfsii. BMC Genomics 11:329
Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU (2003) Dissection of floral induction pathways using global expression analysis. Development 130:6001–6012
Schomburg FM, Patton DA, Meinke DW, Amasino RM (2001) FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13:1427–1436
Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18:1121–1133
Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20:898–912
Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11:445–458
Shimamoto K, Kyozuka J (2002) Rice as a model for comparative genomics of plants. Annu Rev Plant Biol 53:399–419
Shitsukawa N, Ikari C, Shimada S, Kitagawa S, Sakamoto K, Saito H, Ryuto H, Fukunishi N, Abe T, Takumi S, Nasuda S, Murai K (2007) The eikorn wheat (Triticum monococcum) mutant, maintained vegetative phase, is caused by a deletion in the VRN1 gene. Genes Genet Syst 82:167–170
Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003) FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113:777–787
Singh KP, Kushwaha CP (2005) Paradox of leaf phenology: Shorea robusta is a semi-evergreen species in tropical dry deciduous forests in India. Curr Sci 88:1820–1824
Singh KP, Kushwaha CP (2006) Diversity of flowering and fruiting phenology of trees in a tropical deciduous forest in India. Ann Bot 97:265–276
Stinson AJ (2004) Natural selection favors rapid reproductive phenology in Potentilla pulcherrima (Rosaceae) at opposite ends of a subalpine snowmelt gradient. Am J Bot 91:531–539
Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410:1116–1120
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 18:1033–1036
Toth AL, Varala K, Newman TC, Miguez FE, Hutchison SK, Willoughby DA, Simons JF, Egholm M, Hunt JH, Hudson ME, Robinson GE (2007) Wasp gene expression supports an evolutionary link between maternal behavior and eusociality. Science 19:441–444
Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 13:1003–1006
Vasek FC, Sauer RH (1971) Seasonal progression of flowering in Clarkia. Ecology 53:1038–1045
Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, Marden JH (2008) Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol 17:1636–1647
Wang JW, Czech B, Weigel D (2009a) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138:738–749
Wang Z, Gerstein M, Snyder M (2009b) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309:1056–1059
Wilhelm BT, Landry JR (2009) RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48:249–257
Wilkie JD, Sedgley M, Olesen T (2008) Regulation of floral initiation in horticultural trees. J Exp Bot 59:15–28
Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EMN, Maier A, Schwechheimer C (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19:1209–1220
Wilson RN, Heckman JW, Somerville CR (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100:403–408
Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138:750–759
Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D (2009) The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL and APETALA1. Dev Cell 17:268–278
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 19:19581–19586
Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419:308–312
Yasuda J, Matsumoto J, Osada N, Ichikawa S, Kachi N, Tani M, Okuda T, Furukawa A, Rahim Nik A, Nanokaran N (1999) The mechanism of general flowering in Dipterocarpaceae in the Malay Peninsula. J Trop Ecol 15:437–449
Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH (2005) CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol 139:770–778
Zeller G, Henz SR, Widmer CK, Sachsenberg T, Rätsch G, Weigel D, Laubinger S (2009) Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J 58:1068–1082
Acknowledgments
We thank A. Yamaguchi, T. Tsuchimatsu and Y. Takeuchi for helpful discussions and comments on the manuscript, and S. Aikawa, H. Kudoh and A. Satake for valuable discussions through collaboration. This work was supported by grants from the University Research Priority Program in Systems Biology/Functional Genomics of the University of Zurich, SystemsX.ch, and the Swiss National Science Foundation (SNF).
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix 1
Photoperiod pathway
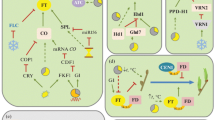
In the photoperiod pathway, flowering is promoted by exposure to inductive photoperiod. Plants sense night length and use it as a flowering cue (reviewed in Imaizumi and Kay 2006; Kobayashi and Weigel 2007). Arabidopsis thaliana is classified as a quantitative long-day plant, meaning that it can flower more rapidly in long-day conditions, while flowering is delayed in short-day conditions. The genes involved in the photoperiod pathway were isolated using mutants defective in day-length-specific acceleration of flowering (Koornneef et al. 1991). In other words, mutants in the photoperiod pathway flower similarly under long-day and short-day conditions. The key components of the photoperiod pathway are the CONSTANS (CO) gene (Putterill et al. 1995) and the FLOWERING LOCUS T (FT) gene. Expression of the CO gene is regulated by circadian clock genes and shows a circadian pattern (Suárez-López et al. 2001; Yanovsky and Kay 2002). In addition, activity of the CO protein is also regulated in a light-dependent manner through the functions of photoreceptors, and thus can induce expression of the downstream gene, FT, only in long-day conditions (Valverde et al. 2004). This provides the crucial mechanism of day length measurement. Expression of the FT gene occurs in leaves, but FT protein moves to the shoot apical meristem (SAM) in which flower development occurs, acting as a ‘florigen’ (Corbesier et al. 2007; Tamaki et al. 2007). Once FT protein arrives at the SAM, it interacts with another protein, FD, and activates downstream floral meristem identity genes such as APETALA 1 (AP1) (Abe et al. 2005; Wigge et al. 2005), and other floral promoters such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) (Michaels et al. 2005; Yoo et al. 2005).
Vernalization pathway
Many individuals of A. thaliana grown in northern Europe need prolonged exposure to the cold of winter to render them competent to flower (Nordborg and Bergelson 1999). This process is called vernalization and ensures that they flower in favorable spring conditions. The vernalization requirement in A. thaliana is conferred by a floral repressor gene, FLOWERING LOCUS C (FLC) (Michaels and Amasino 1999; Sheldon et al. 1999). Without vernalization, highly expressed FLC represses flowering by suppressing the expression of the flowering activators FT, FD and SOC1 (Hepworth et al. 2002; Helliwell et al. 2006; Searle et al. 2006). However, as the plants are exposed to prolonged cold, FLC expression is gradually decreased, and the plants become ready to flower (Michaels and Amasino 1999; Sheldon et al. 1999).
Light quality pathway
Along with the duration of light (photoperiod), light quality is also an important flowering cue. Plants recognize their growing conditions from the light quality, for example, under canopy shade conditions or in the proximity of other plants, and then show acceleration of flowering. This process is mediated by photoreceptors called phytochromes, which function upstream of the FT gene and regulate flowering time (Cerdán and Chory 2003; Halliday et al. 2003).
Thermosensory pathway
Another important cue is ambient temperature. Small changes in ambient temperature regulate flowering time through FT in a thermosensory pathway involving the floral repressor gene, SHORT VEGETATIVE PHASE (SVP) (Lee et al. 2007).
Autonomous pathway
The autonomous pathway is composed of genes that cause late flowering in both long-day and short-day conditions when they are mutated. Therefore, the term “autonomous pathway” is generally used as the pathway that is independent of environmental cues, especially photoperiod. As expected from this definition, the autonomous pathway is not a linear pathway. Rather, it is comprised of subpathways of genes with different biochemical functions (Chan and Struhl 1997; Macknight et al. 1997; Schomburg et al. 2001; He et al. 2003; Simpson et al. 2003; Ausin et al. 2004; Lim et al. 2004; Noh et al. 2004). However, these genes have a common feature in that they control flowering time by mediating the expression level of the floral repressor FLC (Michaels and Amasino 2001).
Gibberellin pathway
Gibberellins (GAs) are a class of plant hormones. In A. thaliana, GAs promote flowering (Wilson et al. 1992; Reeves and Coupland 2001; Willige et al. 2007). GA4 is the most active form of GA in floral induction, and the endogenous levels of GA4 in the SAM are reported to increase up to nearly 100-fold preceding floral initiation (Eriksson et al. 2006). This physiological change in the endogenous GA levels functions through the upregulation of genes such as LEAFY (LFY) and SOC1 (Blázquez and Weigel 2000; Moon et al. 2003).
Age pathway
Plants also sense their age. Plants experience a gradual transition from the juvenile to adult phase as they grow, and this phase transition renders them competent to flower. The abundance of SQUAMOSA PROMOTER BINDING-LIKE (SPL) proteins acts as a key factor in this developmental pathway. A gradual increase in these proteins following the phase transition regulates genes such as FT, SOC1, LFY and AP1 in a direct or indirect manner (Wang et al. 2009a; Wu et al. 2009; Yamaguchi et al. 2009).
Appendix 2
Application of transcriptome profiling to the identification of differentially expressed genes
A central interest in molecular biology has been to identify differentially expressed genes through comparisons of transcription levels before and after specific biological processes. Microarrays have been used widely for this purpose. Microarray platforms provide a reliable, rapid and cost-effective technology with which to analyze genome-wide gene expression patterns by quantifying cDNA by hybridization to probes. This technique has facilitated large-scale transcriptome analysis in many organisms (for example, Schmid et al. 2003; Zeller et al. 2009). However, it has been limited mostly to model organisms with known genome sequences because gene sequences are required to design probes on microarrays.
On the other hand, in nonmodel organisms, expressed sequence tag (EST) sequencing with traditional Sanger capillary sequencers has been used successfully to analyze the transcriptome (for example, Ma et al. 2009; Pirooznia et al. 2010). The EST is a short subsequence of a transcribed cDNA sequence from a known tissue source. The example of strawberry discussed in the main text is one example of EST sequencing (Mouhu et al. 2009). However, this technique is time consuming and costly.
Alternative methods for transcriptomics using high-throughput sequencers have appeared recently (reviewed in Matsumura et al. 2008; Wang et al. 2009b; Wilhelm and Landry 2009). One of the major approaches is RNA-Seq. RNA-Seq provides both the sequence information of transcripts and their expression levels simultaneously. The millions of short reads generated by high-throughput sequencers are either aligned to a reference genome or assembled de novo to infer from which genes they were transcribed. The expression level of each gene can then be measured by counting the number of reads. RNA-Seq has the advantages of higher sensitivity and greater dynamic range of gene expression than microarrays if enough sequencing depth is achieved. Furthermore, it is more rapid and cost-effective than EST sequencing. Owing to advances in methods of de novo assembly, it can be applied to nonmodel organisms with little prior knowledge of their genome sequence (see, e.g., Barakat et al. 2009; Birzele et al. 2010; Ekblom et al. 2010; Rodriguez et al. 2010). RNA-Seq has already been applied to some nonmodel plant species to identify genes expressed differentially in response to several phenomena. For example, several candidate genes responsible for resistance to fungus infections have been revealed through the analyses of differentially expressed genes between fungus-sensitive American chestnut (Castanea dentata) and fungus-resistant Chinese chestnut (Castanea mollissima) (Barakat et al. 2009). As another example, differentially expressed genes in the resurrection plant Craterostigma plantagineum have been characterized during dehydration and rehydration (Rodriguez et al. 2010).
Although transcriptomics using high-throughput sequencers provide useful tools, the high run cost restricts the analysis to a small number of samples. When compared with high-throughput sequencers at the same resolution, microarrays still have an advantage in terms of cost. Therefore, a combination of both microarrays and high-throughput sequencers has been attempted for transcriptomics in nonmodel organisms. Once custom microarrays based on the sequence data generated by high-throughput sequencers have been designed, the microarray serves as a cost-effective tool with which to analyze a large number of samples. There are now some examples of the successful utilization of this system to identify differentially expressed genes (e.g., Bellin et al. 2009; Schmid et al. 2010).
About this article
Cite this article
Kobayashi, M.J., Shimizu, K.K. Challenges in studies on flowering time: interfaces between phenological research and the molecular network of flowering genes. Ecol Res 28, 161–172 (2013). https://doi.org/10.1007/s11284-011-0835-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11284-011-0835-2