Abstract
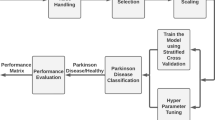
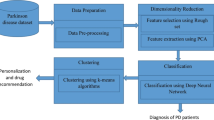
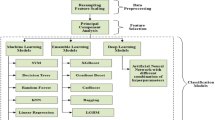
Parkinson’s disease (PD) is a neurodegenerative disorder caused by a deficiency of dopamine in the brain, which is responsible for motor movements. Early detection of PD helps doctors and patients take proper medication and management strategies. Numerous deep learning-based approaches have been developed by researchers in the medical domain to predict PD. However, existing techniques face complications in achieving greater prediction accuracy due to issues such as class imbalance, small dataset size, high false detection rates, overfitting, training complexity, and others. Therefore, this paper proposes a novel automated PD prediction model using an ensemble support-vector deep convolutional-based levy selfish herd (ESDC-LSH) approach. The input dataset consists of data collected from electronic medical records and wearable sensors. To remove data artifacts, preprocessing steps such as missing value filtration, noise removal, duplicate record elimination, and normalization are performed. After preprocessing, features with valuable information are selected and weighted based on their significance. Using the proposed ESDC-LSH approach, healthy and PD-infected subjects are accurately classified. Additionally, the proposed approach further categorizes diseased subjects based on illness levels, categorized into low, medium, and high risk. Based on the evaluated risk levels, healthcare professionals can use an ontology-based recommendation system to provide suitable medication for the patients. The effectiveness of the proposed ESDC-LSH technique is examined by evaluating measures such as accuracy, precision, Root Mean Square Error (RMSE), sensitivity, F1-score, specificity, and Mean Absolute Error (MAE). Simulation outcomes demonstrate that the proposed ESDC-LSH approach achieves higher prediction accuracy, about 98.8%, precision of about 98.48%, RMSE of about 0.198, sensitivity of about 97.25%, F1-score of about 97.61%, specificity of about 97.45%, and MAE about 0.10 compared to other techniques.










Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Wang, W., Lee, J., Harrou, F., & Sun, Y. (2020). Early detection of Parkinson’s disease using deep learning and machine learning. IEEE Access, 8, 147635–147646.
Devos, D., Cabantchik, Z. I., Moreau, C., Danel, V., Mahoney-Sanchez, L., Bouchaoui, H., Gouel, F., Rolland, A. S., Duce, J. A., & Devedjian, J. C. (2020). Conservative iron chelation for neurodegenerative diseases such as Parkinson’s disease and amyotrophic lateral sclerosis. Journal of Neural Transmission, 127(2), 189–203.
Torrisi, M., Pollastri, G., & Le, Q. (2020). Deep learning methods in protein structure prediction. Computational and Structural Biotechnology Journal, 18, 1301–1310.
Choudhury, A., & Mukherjee, S. (2020). In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. Journal of Medical Virology, 92(10), 2105–2113.
Khan, A., Khan, M., Saleem, S., Babar, Z., Ali, A., Khan, A. A., Sardar, Z., Hamayun, F., Ali, S. S., & Wei, D. Q. (2020). Phylogenetic analysis and structural perspectives of RNA-dependent RNA-polymerase inhibition from SARs-CoV-2 with natural products. Interdisciplinary Sciences: Computational Life Sciences, 12(3), 335–348.
Sanavia, T., Birolo, G., Montanucci, L., Turina, P., Capriotti, E., & Fariselli, P. (2020). Limitations and challenges in protein stability prediction upon genome variations: Towards future applications in precision medicine. Computational and Structural Biotechnology Journal, 18, 1968–1979.
Tracy, J. M., Özkanca, Y., Atkins, D. C., & Ghomi, R. H. (2020). Investigating voice as a biomarker: Deep phenotyping methods for early detection of Parkinson’s disease. Journal of Biomedical Informatics, 104, 103362.
Soumaya, Z., Taoufiq, B. D., Benayad, N., Yunus, K., & Abdelkrim, A. (2021). The detection of Parkinson disease using the genetic algorithm and SVM classifier. Applied Acoustics, 171, 107528.
Alford, R. F., Fleming, P. J., Fleming, K. G., & Gray, J. J. (2020). Protein structure prediction and design in a biologically realistic implicit membrane. Biophysical Journal, 118(8), 2042–2055.
Ovsyannikova, I. G., Haralambieva, I. H., Crooke, S. N., Poland, G. A., & Kennedy, R. B. (2020). The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunological Reviews, 296(1), 205–219.
Ye, C., Li, J., Hao, S., Liu, M., Jin, H., Zheng, L., Xia, M., Jin, B., Zhu, C., Alfreds, S. T., & Stearns, F. (2020). Identification of elders at higher risk for fall with statewide electronic health records and a machine learning algorithm. International Journal of Medical Informatics, 137, 104105.
Talitckii, A., Kovalenko, E., Anikina, A., Zimniakova, O., Semenov, M., Bril, E., Shcherbak, A., Dylov, D. V., & Somov, A. (2020). Avoiding misdiagnosis of Parkinson’s disease with the use of wearable sensors and artificial intelligence. IEEE Sensors Journal, 21(3), 3738–3747.
Masud, M., Singh, P., Gaba, G. S., Kaur, A., Alroobaea, R., Alrashoud, M., & Alqahtani, S. A. (2021). CROWD: Crow search and deep learning based feature extractor for classification of Parkinson’s disease. ACM Transactions on Internet Technology (TOIT), 21(3), 1–18.
Sharma, P., Sundaram, S., Sharma, M., Sharma, A., & Gupta, D. (2019). Diagnosis of Parkinson’s disease using modified grey wolf optimization. Cognitive Systems Research, 54, 100–115.
Divagar, M., Gayathri, R., Rasool, R., Shamlee, J. K., Bhatia, H., Satija, J., & Sai, V. V. R. (2021). Plasmonic fiberoptic absorbance biosensor (P-FAB) for rapid detection of SARS-CoV-2 nucleocapsid protein. IEEE Sensors Journal, 21(20), 22758–22766.
Kaur, S., Aggarwal, H., & Rani, R. (2020). Hyper-parameter optimization of deep learning model for prediction of Parkinson’s disease. Machine Vision and Applications, 31(5), 1–15.
Bahaddad, A. A., Ragab, M., Ashary, E. B., & Khalil, E. M. (2022). Metaheuristics with deep learning-enabled Parkinson’s disease diagnosis and classification model. Journal of Healthcare Engineering, 2022, 1–14.
Ali, F., El-Sappagh, S., Islam, S. R., Kwak, D., Ali, A., Imran, M., & Kwak, K. S. (2020). A smart healthcare monitoring system for heart disease prediction based on ensemble deep learning and feature fusion. Information Fusion, 63, 208–222.
Rahman, T., Chowdhury, M. E., Khandakar, A., Islam, K. R., Islam, K. F., Mahbub, Z. B., Kadir, M. A., & Kashem, S. (2020). Transfer learning with deep convolutional neural network (CNN) for pneumonia detection using chest X-ray. Applied Sciences, 10(9), 3233.
Sørensen, L., Nielsen, M., & Alzheimer’s Disease Neuroimaging Initiative. (2018). Ensemble supports vector machine classification of dementia using structural MRI and mini-mental state examination. Journal of Neuroscience Methods, 302, 66–74.
Zhao, R., Wang, Y., Liu, C., Hu, P., Li, Y., Li, H., & Yuan, C. (2020). Selfish herd optimizer with levy-flight distribution strategy for a global optimization problem. Physica A: Statistical Mechanics and its Applications, 538, 122687.
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have not disclosed any competing interests.
Human and Animal Rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Caroline El Fiorenza, J., Sellam, V. ESDC-LSH: Ensemble Support-Vector Deep Convolutional Based Levy Selfish Herd Optimization for Prediction and Classification of Parkinson’s Disease. Wireless Pers Commun 135, 1861–1883 (2024). https://doi.org/10.1007/s11277-024-11173-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11277-024-11173-5