Abstract
In this paper, a series of processes are performed to assess the degree of infection in a subject’s blood infected with malaria. This process improves the existing testing method’s inefficiencies to assess the results obtained of the infected blood. Image processing algorithm was constructed to establish a standardized test procedure as a way to process the information. The algorithm is built to process the images obtained from the microscopic image of the infected blood and detect the extent of Plasmodium infection which is the cause of Malaria Disease. The images obtained from the infected red blood cells and uninfected red blood cells are processed using the image processing algorithm to distinguish the number of cells infected by malaria in the infected blood. A quantitative analysis is carried out using the results to evaluate the extent of the infection in the blood cells. To establish the relationship between the number of infected and uninfected cells, the algorithm was constructed based on color image processing through HSV color space and morphological image processing techniques. The key configuration for the proposed algorithm is the morphological image processing technique which clearly divide the red blood cells in the micrograph based on the extent of penetration of plasmodium in the blood cells using the clamp splitting method and categorize the infected blood cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Malaria is the symptom of acute febrile infection. It is caused because the plasmodium parasite of red blood cells [1]. About 3.2 billion people remain at risk of malaria. In 2015 alone, there were an estimated 214 million new cases of malaria and 438,000 deaths [2]. Millions of people are still not accessing the services they need to prevent and treat malaria [2]. Depending on the type of plasmodium, malaria treatment is different. To reduce the progression of infection, a fast and accurate diagnosis at an early stage is very important [3, 4]. Until now, the diagnosis of malaria is carried out using the Giemsa staining the infected blood and a standard test procedure is followed to read the thin smear and the thick smear on the obtained microscopic image. Though this method is relatively economical and detects the possible parasites and the distinction of species, the results are subjective and need high skills [5]. But this method is inadequate because of complex, labor-intensive and long duration tests.
Therefore, the image processing algorithm is developed as a way of establishing the standardized inspection procedure in order to determine the extent of malaria infection. This process is derived as an alternate procedure to improve the inefficiency of conventional testing method. The present study was carried out through the microscope image of the blood infected with malaria. The degree of infection was determined by detecting the plasmodium that causes malaria. The series of processes includes the procedure to check the quantity plasmodium in each of red blood cells (RBCs). The proposed algorithm is composed of morphological image processing technique to clearly divide the RBCs in the micrograph which are infected and then using clamp splitting method and HSV color image processing to categorize them as infected.
2 Parasitemia Evaluation Methods
Evaluation algorithm for detecting the degree of blood infection, using MATLAB software, was designed with respect to plasmodium infected red blood cells (RBCs) images. This process enables the algorithm to distinguish between infected and uninfected RBCs images. In order to determine parasitemia values, the proposed algorithm is composed of morphological image processing technique to effectively identify the infected and uninfected RBCs. The algorithm also detects the plasmodium penetration into the RBCs which is a key configuration in the series of processes. The algorithm is made as a standardized test procedure through color image processing based on the HSV color space technique providing a quantitative analysis on the blood samples.
2.1 Preprocessing
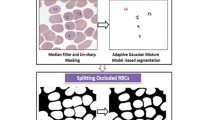
RBCs images have some preprocessing steps which involves the conversion of the original image into gray scale image and the enhancement its contrast by adaptive histogram equalization [6, 7, 8]. The value of adaptive histogram equalization was determined by Otsu’s method. Through these two processes an improved image is obtained for the next stage. Figure 1 shows a prior image and subsequent image with respect to the preprocessing. Figure 1b shows the grey scale image of the actual microscopic image which categorically distinguishes the RBCs in the sample (Fig. 2).
2.2 Edge Detection
The Sobel edge detector uses two 3 × 3 kernels which are convolved with the original image to calculate approximations of the derivatives, one for horizontal changes, and one for vertical.
Edge detection is a general term of the mathematical method to determine the point in the digital image which detects the change in the brightness of the image if it changes rapidly or having a discontinuity.
Edge refers to a point which is composed of a set of typical curve segment to radically change the brightness of the image. Therefore, applying the image edge detection algorithm can reduce the amount of data to be handled markedly, while maintaining the important structural characteristics of the image and filter the lower priority information.
The Sobel edge detector (SED) is a discrete differentiation operator, computing an approximation of the gradient of the image intensity function. At each point in the image, the result of SED is either the corresponding gradient vector or the norm of this vector. SED uses the masks in Fig. 3 to approximate digitally the first derivatives Gx and Gy. In other words, the gradient at the center point in a neighborhood is computed as follows by SED [9, 10].
A pixel at location \( \left( {x, y} \right) \) is an edge pixel if \( {\text{g }} \ge {\text{T}} \) at that location, where T is a specified threshold.
2.3 Edge Link
If the edge detection procedure is successful, the subsequent operations for interpreting the contents of the image information can be substantially simplified. However, it is not possible to obtain an ideal edge, such as the actual image of the appropriate complexity. For this reason, it may represent a potential corresponding to the discontinuity of the image brightness. Edges are extracted from the non-trivial image, which do not complicate the subsequent operations to interpret the image data corresponding to the respective conditions of interest in the image. This means that not only the edge segments are connected by a curved edge as false edges or omissions. Edge detection is a technique of the basic steps in a variety of fields such as image processing, image analysis, image recognition, computer vision. Therefore, the edge link has been recognized as an essential step in the image processing procedure.
Derived edges are connected to each other at their link points that forms closed perimeter around the RBCs. Link points are close the shape of edge lines using 20 kinds of different 3 × 3 masks shown in Fig. 4. Such edge link mask is connected to each link of all points through the scanning area with respect to the image size of 1300 × 1030.
2.4 Morphological Operation
For RBCs images segmentation, the morphological operation was composed of following 3 sequential steps: (1) flood-fill operation, (2) remove invalid RBCs and noise, (3) detect overlap RBCs. As a prerequisite for the processing of the following steps, the following Fig. 5 shows the result of each step in the morphological operation.
First, based on the edge image linked, a flood-fill operation on background pixels (that is internal pixels of the linked edge contour) of the input binary image was performed. However, due to the presence of one linked edge contour inside another linked edge contour, the flood-fill operation is not performed accurately. In other words, a black cavity occurs in the image of the white colored RBCs in which the flood-fill operation is performed. Opening and closing operation was carried out to improve this phenomenon. Opening generally smoothen the contour of an object, breaks narrows isthmuses, and eliminates thin protrusions. Closing also tends to smooth sections of contours but, as opposed to opening, it generally fuses narrow breaks and long thin gulfs, eliminates small holes, and fills gaps in the contour [11, 12].
The opening of set A by structuring element B, denoted A ∘ B, is defined as
Thus, the opening A by B is the erosion of A by B, followed by a dilation of the result by B. Similarly, the closing of set A by structuring element B, denoted A · B, is defined as
It means that the closing of A by B is simply the dilation of A by B, followed by the erosion of the result by B.
Second, in order to remove invalid RBCs and the image noise the following two methods are used. One is the method for removing the edge line contacting the outermost pixel on the image that is to say the removal of invalid RBCs creates the outermost pixel line of the image. It connects the outermost pixel line to the adjacent linked edge line. Then it removes one connected line with the image coordinates of the outermost pixel of the image, the other is the method for removing objects smaller than the size of typical RBCs. In the process of removing noise from the image, the size of the smallest RBC is calculated based on the number of pixels, and the largest noise is calculated. The threshold value is set by the two calculated sizes.
Third, labeling was performed as the first step to detect the overlap RBCs. As a result of labeling, all RBCs are merged into one object with or without overlapping. The objects which were rolled into one have morphological differences. Identification of distorted values based on the roundness of each object is a criterion of distinguishing the overlap RBCs. Therefore the reason for performing the labeling is that it is possible for the roundness numerically confirmed. Roundness of overlap RBCs is close to zero, the roundness of the correct circle is expressed as 1, since the roundness means a correct circle. In this research, the RBCs were determined to overlap less than 0.6, because the threshold value of the roundness was set to 0.6 (Figs. 6, 7).
2.5 Clump Split
The purpose to enforce Clump split overlap on RBCs is to improve significantly the accuracy in the evaluation process of parasitemia. That is, as shown in Fig. 8, it arises a problem that even the overlap of two RBC recognized as one RBC. Then, the clump split method [13, 14, 15] is used to split clumps of two or more RBCs into constituent cells. For the contents of this process, the process is shown through the following in Fig. 8 and has been explained based on it. Processed results by the clump split method is shown in Fig. 9.
Concavity depth \( \overline{CD} \) is a concaveness measure proposed by Rosenfeld [16]. First the group \( K_{i} \) of convex hull segment can be obtained from image boundary. Then calculating the maximum distance between each segment it can be acquired for four pixels, there are composed of \( K_{1} , K_{2} , K_{3} , K_{4} \). Using these four pixels may draw the two tangents \( \overline{{K_{1} K_{3} }} \;{\text{and}}\;\overline{{K_{2} K_{4} }} \). Each pixel on the boundary between \( K_{1} \) and \( K_{3} \) has a concavity depth value, which is the rectilinear distance from the corresponding tangents \( \overline{{K_{1} K_{3} }} \). Where the distance from \( K_{1} \) to \( C_{i} \) is a value that can be seen by the calculation. This calculation process is continued until the calculated maximum value of the concavity depth \( \overline{{CD_{1} }} \). Also each pixel on the boundary between \( K_{2} \) and \( K_{4} \) has a concavity depth value, which is the rectilinear distance from the corresponding tangents \( \overline{{K_{2} K_{4} }} \). Where the distance from \( K_{2} \) to \( C_{i} \) is a value that can be seen by the calculation. This calculation process is continued until the calculated maximum value of the concavity depth \( \overline{{CD_{2} }} \). After performing this iterative process, finally, it turns the value of the closest pixel in clump split line \( \overline{{C_{i} C_{i + n} }} \).
2.6 Parasites Detection
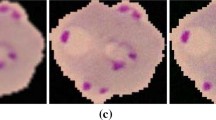
To detect malaria parasites and the method using the Giemsa stain, Plasmodium is stained with a purple throughout the process (Fig. 10). In order to extract these characteristics through the Figs. 11 and 12 compare the RGB image and HSV image. The results were excellent in the HSV image.
Hue, saturation, value (HSV) is one of several color system used by people to select colors (e.g., of paints or inks) from a color wheel or palette. These color systems is very closer than the RGB system to humans experience and expression about color sensations [9, 17,18,19].
For parasites detection, it can be divided following 4 sequential steps: (1) HSV conversion, (2) S component analysis, (3) parasites detection, (4) image registration.
In a color image, the sub-images in red, green, and blue are very analogous to each other. The separation of parasites in gray levels is not proper. The parasites were found to be different in color. Therefore, hue–saturation–value color space is analyzed as shown in Fig. 12. A bright object is a viewpoint in hue component; H and saturation; S. This is because Giemsa staining solution colors hexane. That is, RGB colors emphasized in these objects look dark and are colored in purple [20, 21]. Therefore, the white cells and parasites that include RGB are much brighter in images than other objects. This is why saturation components are used in color images (Fig. 13).
3 Experimental Results
Figure 14 shows the parasites evaluation results obtained using the MATLAB tool. The microscopic image of the blood sample is processed through various preprocessing, processing and post processing stages. Evaluation algorithm for blood infection degree, using MATLAB software, were designed with respect to red blood cells (RBCs) image infected with parasites of the plasmodium to distinguish uninfected RBCs image. Briefly, this involved the detection of the infected RBC using preprocessing (which involves the conversion of gray scale and the enhancement of the image contrast by adaptive histogram equalization (Fig. 15).
The value of adaptive histogram equalization was determined by Otsu’s method), edge detection (It uses two 3 × 3 kernels which are convolved with the original image to calculate approximations of the derivatives, one for horizontal changes, and one for vertical), edge link (Link points are close the shape of edge lines using 20 kinds of different 3 × 3 masks. Such edge link mask is connected to each link of all points through the scanning area with respect to the image size of 1300 × 1030), morphological operation (It was composed of flood-fill operation, remove invalid RBCs and noise, detect overlap RBCs), clump split (Which is used to split clumps of two or more RBCs into constituent cells), parasites evaluation (it can be divided HSV conversion, S component analysis, parasites detection, image registration.). Subsequently, parasite infection is identified based on the S component analysis of converted HSV image. These color systems are very closer than the RGB system to humans experience and expression about color sensations. It was found that all these extensive steps have improved processing of the image and assisted in evaluating the number of cells that are infected with malaria. When compared to the existing process to identify malaria infection, this procedure is fast, accurate, economical and does not require high skills to interpret the results. The interface tool is designed in such a way that the sequencing is given in the order in which the procedure should be executed and therefore reduces the ambiguity in evaluating the results. The result table gives the RBC count, number of infected RBC and % of parasitemia present in the blood sample. The results also indicate that that the method is robust and accurate.
4 Conclusions
-
1.
In order to determine the extent of malaria infection, the image processing algorithm was developed as a way of establishing the standardized inspection procedures.
-
2.
This process is derived and resulted in the improvement to the inefficiency of conventional testing method.
-
3.
The present study was carried out through the microscope image of the blood infected with malaria, to help determine the degree of infection by detecting the plasmodium that causes malaria.
-
4.
The series of processes are procedural connected to check the quantity of each of red blood cells (RBCs).
-
5.
The proposed algorithm is composed of morphological image processing to clearly divide the RBCs in the micrograph of infected RBCs and clamp splitting method, also to detect a plasmodium penetration into RBCs.
-
6.
It was constructed as a standardized test procedure through color image processing based on the HSV color space and morphological image processing techniques.
-
7.
The results obtained are robust with an efficiency of 40% over the conventional techniques in terms of speed and accuracy.
Change history
05 October 2017
The Acknowledgements section in the original publication is incorrect. The correct Acknowledgements section is given here.
References
Sinden, R., & Gilles, H. (2002). The malaria parasites. In D. A. Warrell & H. M. Gilles (Eds.), Essential malariology (4th ed.). London: Arnold.
WHO Global Malaria Programme. (2015). Section 2: Trends in infection prevalence, cases and deaths. World Malaria Report 2015 (pp. iv–v). Geneva: WHO.
WHO (Ed.) (2010). Guidelines for the treatment of malaria (2nd ed., pp. 1–10). Geneva: WHO.
Ross, N. E., Pritchard, C. J., Rubin, D. M., & Dusé, A. G. (2006). Automated image processing method for the diagnosis and classification of malaria in thin blood smears. Medical and Biological Engineering and Computing, 44(5), 427–436.
Amexo, M., Tolhurst, R., Barnish, G., & Bates, I. (2004). Malaria misdiagnosis: Effects on the poor and vulnerable. The Lancet, 364(9448), 1896–1898.
Karel, Z. (1994). Contrast limited adaptive histogram equalization. Graphics gems IV (pp. 474–485). (code: 479–484).
Reza, A. M. (2004). Realization of the contrast limited adaptive histogram equalization (CLAHE) for real-time image enhancement. VLSI Signal Processing, 38(1), 35–44.
Cheng, H. D., & Shi, X. J. (2004). A simple and effective histogram equalization approach to image enhancement. Digital Signal Processing, 14(2), 158–170.
Gonzalez, R. C., Woods, R. E., & Eddins, S. L. (2011). Digital image processing using MATLAB (2nd ed.). Singapore: McGraw Hill.
Vincent, O. R., & Folorunso, O. (2009). A descriptive algorithm for sobel image edge detection. In Proceedings of informing science and IT education conference (pp. 97–107).
Gonzalez, R. C., & Woods, R. E. (2010). Digital image processing (3rd ed.). New Jersey: Pearson.
Goyal, M. (2011). Morphological image processing. International Journal of Computer Science and Technology, 2(2), 161–165.
Kumar, S., Ong, S. H., Ranganath, S., Ong, T. C., & Chew, F. T. (2005). A rule-based approach for robust clump splitting. Pattern Recognition, 39(6), 1088–1098.
LaTorre, A., Alonso-Nanclares, L., Muelas, S., Peña, J. M., & DeFelipe, J. (2013). Segmentation of neuronal nuclei based on clump splitting and a two-step binarization of images. Expert Systems with Applications, 40, 6521–6530.
Farhan, M., Yli-Harja, O., & Niemistö, A. (2013). A novel method for splitting clumps of convex objects incorporating image intensity and using rectangular window-based concavity point-pair search. Pattern Recognition, 46(3), 741–751.
Rosenfeld, A. (1985). Measuring the sizes of concavities. Pattern Recognition Letters, 3(1), 71–75.
Georgieva, L., Dimitrova, T., & Angelov, N. (2005). RGB and HSV colour models in colour identification of digital traumas images. In International conference on computer systems and technologies (pp. V.12.1–V.12.6).
Sural, S., Qian, G., & Pramanik, S. (2002). Segmentation and histogram generation using the HSV color space for image retrieval. In IEEE ICIP (pp. II.589–592).
Deswal, M., & Sharma, N. (2014). A fast HSV image color and texture detection and image conversion algorithm. International Journal of Science and Research, 3(3), 1279–1284.
Harms, H., Gunzer, U., & Aus, H. M. (1986). Combined local color and texture analysis of stained cells. Computer Vision, Graphics and Image Processing, 33(3), 364–376.
Olivo-Marin, J. C. (2002). Extraction of spots in biological images using multiscale products. Pattern Recognition, 35(9), 1989–1996.
Acknowledgements
This research was supported by basic science research program through the National Research Foundation of Korea (NRF) funded by the ministry of education, Science and technology (No. 2013R1A1A2061581) and financially supported by the National Foundation of Korea (NRF) funded by the Korea Government (MISP) (No. 2014R1A2A1A11053533).It was also supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. 2016R1A6A1A03012069).
Author information
Authors and Affiliations
Corresponding author
Additional information
A correction to this article is available online at https://doi.org/10.1007/s11277-017-4974-9.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kim, CH., Kwac, LK. & Kim, HG. The Development of Evaluation Algorithm for Blood Infection Degree. Wireless Pers Commun 98, 3129–3144 (2018). https://doi.org/10.1007/s11277-017-4136-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11277-017-4136-0