Abstract
Onion skins, actually recycled as organic fertilizers, could be used as a substrate in environmental-friendly bioprocesses to recover high-value bioactive compounds and food ingredients.
In this work, a bioprospecting method was carried out including 94 bacterial and 45 yeast strains from several agri-food and environmental niches to verify their ability to grow on onion skins as unique nutrients source.
Red and yellow onion skins were assessed by newly selected starter-driven liquid submerged fermentation assays mainly aimed at the release and modification of polyphenols through microbial activities. Fermented onion skins were also investigated as a inexpensive favourable source of microbial enzymes (amylases, proteases, lipases, esterases, cellulases, xylanases).
In red onion skins, the treatment with Lactiplantibacillus plantarum TB 11–32 produced a slight increase of bioactive compounds in terms of total phenolics, whereas with the yeast strain Zygosaccharomyces mrakii CL 30 − 29 the quercetin aglycone content increased of about 25% of the initial raw material.
In yellow onion skins inoculated, the highest content of phenolic compounds was detected with the yeast strain Saccharomyces cerevisiae En SC, while quercetin aglycone increased of about 60% of the initial raw material in presence of the bacterial strain L. plantarum C 180 − 34.
In conclusion, the proposed microbial pre-treatment method can be a potential strategy to re-use onion skins as a fermentation substrate, and as a first step in the development of a biorefinery process to produce value-added products from onion by-products.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The world production of onion (Allium cepa L.) is about 47 million tons every year and, during the past 20 years, this horticultural crop became the second most important one (FAOSTAT 2018). Before harvest, onion bulbs are generally pulled from the soil to stop growth. Then, they are cured for a few days in the field to reduce excess moisture from the outer skin and neck to prevent excessive shrinkage of the onion. During storage this step allows also for color development. A huge amount of waste (stalk, roots and skins) derives from industrial processing of onions. In Europe, a by-product production of about 50,000 tons per year was registered by Spain, Netherlands, and the UK, the main European onion producers (Roldan et al., 2008; Sharma et al. 2016; Santiago et al. 2020).
The onion wastes are rich in fiber and bioactive compounds and represent a source to develop different bioproducts such as polyphenols, pigments, organic acids, enzymes, and bioenergy (Sagar and Pareek 2020). However, the different chemical composition of onion waste categories limits their use as whole waste.
Onion skins removed from edible bulbs represent the major waste of onion processing, since they are unsuitable as landfill disposal or animal feed because of their composition and smell. Therefore, sustainable onion production requires the valorization of their skins by turning this waste into “by-product” (Gontard et al. 2018). Onion skin waste is a source of phenolic chemicals, primarily quercetin and its derivatives, luteolin, and kaempferol, myricetin, isorhamnetin derivatives and carbohydrates (Benito-Román et al. 2020; Sagar et al. 2020). Onion skin cell walls show a very complex structure, mainly formed by pectin (42.4%), hemicelluloses (36.6%), and cellulose (21%) (Munir et al. 2018). This by-product is also an interesting source of pectin, which is highly requested at global level (Kim et al. 2019) and the phenolic compounds can have many pharmacological properties, such as antioxidant, antimicrobial, anticarcinogenic, antimutagenic activities, and their antidiabetic potential (Benitez et al., 2011; Ren and Zhou 2021).
The theory of waste valorization is strictly associated with sustainable technologies for the recycling and the reuse. Fermentation is one of the most promising frontiers to start from biomass feedstock and, through various techniques such as solid-state fermentation (SSF), submerged fermentation (SmF), or anaerobic digestion to recover the nutrients by a biological extraction process, by creating value-added products and “green” energy (Carmona-Cabello et al. 2018).
Whole microbial cells can act on the cell wall matrix, leading to the bioconversion of the plant fibers i.e. cellulose, hemicellulose, pectin and lignin, and allowing the release of polyphenols and organic acids (Barcelos et al. 2020; Ramires et al. 2020; Sayago-Ayerdi et al., 2021). Several studies demonstrated that during fermentation of agri-food wastes microbial hydrolitic enzymes, such as amylase, protease, β-glucosidase, xylanase, cellulase can break down cell wall components and consequently facilitate phenolic mobilization, whereas esterase and lipase can be responsible of phenolics evolution essentially by esterification (Bei et al. 2018; Dey et al. 2016; Hur et al. 2014; Yin et al. 2020). The predominance of one or more of these enzymatic activities, positively correlated with the increase of the release or the bioavailability of phenolic compounds, can be considered a key attribute for the selection of new candidate microbial starter for the treatment of agri-food by-products.
The fermentation process has been considered as an efficient method to obtain the chemical modification of phenolic compounds from agro-industral by-products, and to enhance their release and eventually to increase the antioxidant activity (Gulsunoglu-Konuskan and Kilic-Akyilmaz 2022). Improvement of bioactive compounds extraction from plant products using a microbial fermentation process has been reported before, e.g., for apple pomace, black soybeans and Larrea tridentate leaves olive mill wastewater, orange skin waste (Cheng et al. 2013; Ramires et al. 2020). In SmF, the microorganisms develop in a liquid medium obtaining the nutrients required for their growth and metabolic activities by the raw material. This process allows the monitoring of the relevant parameters for microbial growth, such as pH and temperature, with very high efficiency.
Several fermentation approaches were explored for potential re-using onion wastes by lignocellulosic biomass treatment, such as for bioethanol production in presence (Kim et al. 2019) or not (Ganguly et al. 2021) of a enzymatic saccharification step for bio-sugar production and for producing acetone, butanol, ethanol using Clostridium culture (Poe et al. 2020). Also, acetic acid was obtained by fermenting dried onion waste with Acetobacter acetii (Kim et al. 2019).
In this study, two different onion cultivars, the red onion cv “Cipolla rossa di Tropea” and the yellow onion cv Recas were chosen, since they are common in commercial productions in Italy and Spain, respectively. The traditional and commercially relevant Italian Protected Geographic Indication (PGI) onion variety “Rossa di Tropea” from Calabria Region (Italy) is an outstanding food product with several peculiar organoleptic properties and a distinctive taste made by a delicate and persisting aroma. The yellow cv Recas is late cycle and long-day onion, with a good firmness and high density and good storage capacity. Benitez et al. (2011) reported that the skins, which are rich in polyphenols and flavonoids and a relative high antioxidant activity, could be considered as a potential functional ingredient. Moreover, the remarkable bioactivity of red onion “Cipolla rossa di Tropea” skins suggested several potential applications in different industrial sectors as nutraceuticals, pharmaceuticals and food (Celano et al. 2021).
The main objective of this study is to valorize onion skins by using a microbiological treatment able to improve the release and the extraction of polyphenols content of these by-products.
In this study microorganisms (yeasts and bacteria) from different natural origins were selected for their ability to grow in presence of red and yellow onion skins as unique nutrient sources.
SmF approach on red and yellow onion skins, inoculated with selected microbial starters, was applied to enhance the release of polyphenols as well as on changes in their profile and relative amount of single polyphenols. By this approach, an improved recovery of polyphenols from the onion skins and an enrichment in the aglyconic form of specific phenolic compounds, are expected. The hypothesis is that microorganisms can (i) act by hydrolytic enzymes on the vegetable cell wall structure and glycosidic linkages, releasing polyphenols and also (ii) use their secondary metabolism to promote the chemical modification of phenolic compounds (Slama et al. 2021; Gulsunoglu-Konuskan et al. 2021). Following this route, the major microbial enzymatic activities found in the fermented products were also investigated.
Materials and methods
Feedstock sampling
Red onion skins (cv Cipolla rossa di Tropea) were kindly supplied by the Italian company Azienda Agricola Veltri srl (CZ, Italy), yellow onion skin samples (cv Recas variety Citation) were provided by PROCECAM (Association of Onion Producers of Castilla-La Mancha, Spain). Onion skin samples were packed in aerated bags at production plant sites in June 2021 and transferred to the laboratory in 3–5 days, then samples were immediately grinded by a laboratory knife mill (Grindomix GM 200, RETSCH Gmbh, Haan, Germany) at maximum speed for 5 min and stored at – 20 °C for further experiments.
Microbial species and culture conditions
In this study, a number of 94 bacterial strains were chosen among two phyla frequently associated with plant material and the production of fermented foods:
-
Proteobacteria (alphaproteobacteria Acetobacter, Agrobacterium, Sphingomonas; gammaproteobacteria Dickeya, Enterobacter, Erwinia, Hafnia, Pantoea, Pectobacterium, Pseudomonas, Serratia);
-
Bacillota (synonym Firmicutes, Oren and Garrity 2021) (Bacillus, Lactiplantibacillus, Leuconostoc, Levilactobacillus, Paenibacillus, Pediococcus, Staphylococcus).
Also, a number of 45 yeast strains were selected among species (Aureobasidium, Candida, Debaryomyces, Geotrichum, Hanseniaspora, Kluyveromyces, Metschnikowia, Pichia, Rhodotorula, Saccharomyces, Trichosporon, Zygosaccharomyces) generally associated to food fermentation and treatment of waste/by-products of vegetable origin.
The entire collection set of yeast and bacterial strains were selected from the CNR-ISPA microbial collection (Table S1 and S2).
Bacillus spp. strains were grown on Nutrient agar medium (Merck KGaA, Darmstadt, Germany) at 25 °C for 2–3 days; Bacillus simplex strains, Paenibacillus spp., Sphingomonas spp. were grown on Tryptic soy agar (Merck KGaA, Darmstadt, Germany) at 30 °C for 2–3 days; LAB strains were grown on Man, Rogosa and Sharpe (MRS) agar medium (Microbiol Diagnostics, Cagliari, Italy) at 30 °C for 3–5 days under anaerobic conditions; Staphylococcus spp. were grown on MRS Agar with replacement of glucose as carbon source with sucrose and addition of 7.5% marine salts (VibrantSea, Seachem Laboratories Inc., Madison GA, USA) at 37 °C for 3–5 days, under anaerobic conditions. Pseudomonas and Enterobacteriaceae strains were grown on Nutrient Broth added with 1% (w/v) glucose at 30 °C for 1–2 days. Acetobacter spp. were grown on GYC medium (Hommel 2014) modified as follows (mGYC): 50 g/L glucose (VWR Chemicals, Leuven, Belgium), 10 g/L yeast extract (Merck KGaA, Darmstadt, Germany), 2 g/L CaCO3 (Merck KGaA, Darmstadt, Germany) for 48–72 h at 28 °C. All media were added with 0.05 g/L nystatin.
Yeast strains were all grown on Sabouraud dextrose agar medium (LABM, Heywood, Lancashire, UK) added with 0.1 g/L of ampicillin (Sigma-Aldrich, Darmstadt, Germany) and 0.05 g/L of kanamycin (Sigma-Aldrich, Darmstadt, Germany) and incubated at 30 °C for 2–4 days.
Yeast and bacteria collection screening on onion skins-based media
Microbial media for yeast and bacteria collection screening were prepared following the indications of Niu et al. (2017) with some modifications: 2 g/L casein peptone (Sigma-Aldrich, Darmstadt, Germany), 2 g/L soy peptone (Sigma-Aldrich, Darmstadt, Germany), 2 g/L NaCl (Sigma-Aldrich, Darmstadt, Germany), 20 g/L agar (Sigma-Aldrich, Darmstadt, Germany) and a specific percentage (w/v) of ground onion skins. Preliminary experiments have been performed in order to evaluate the suitable quantity of the onion skins to prepare the agar media (data not shown). The final selected percentages of the ground onion skins added to the mixture were 3-4-5% (w/v). These concentrations are corresponding to a final hydrated ground onion skins preparation percentage of 15-20-25% (w/v). The weight increase of hydrated onion skins was measured after the incubation of the initial samples in presence of water excess for 30 min and the draining until the weight was stable. It resulted about 5-fold higher than the initial weight value.
The incubation conditions were the same used for the growth on laboratory media. Microbial strains were screened for their ability to survive and grow on different onion skins (OS) concentrations by applying an arbitrary scale: 0, no growth; 1, slight growth; 2, medium growth; 3, intense growth (similar to the growth optimal medium, used as control); 4, > growth on the optimal medium.
The growth scores were used to obtain a Weighted Mean Growth (WMG) value for each strain. The equation was defined as follows:
WMG = [(N1 × 2) + (N2 × 3) + (N3 × 5)]/10.
N1 = scale growth on 15% concentration OS.
N2 = scale growth on 20% concentration OS.
N3 = scale growth on 25% concentration OS.
2 = multiplier value for the growth at 15% concentration OS.
3 = multiplier value for the growth at 20% concentration OS.
5 = multiplier value for the growth at 25% concentration OS.
10 = numerator (sum of the multiplier values).
Laboratory-scale fermentation assays
A submerged batch fermentation was set up for feedstocks treatment. The raw material was homogenized and used to prepare samples to be singularly inoculated with selected microbial strains. The final volume fermentation in glass jars (100 mL) was prepared adding 25 g of hydrated onion skins and 50 mL drinking water. Prepared mix was heat treated in water bath at 90 °C for 10 min, cooled and inoculated with 25 mL of fresh microbial suspension prepared as follows. Firstly, microbial inoculum was prepared growing the selected strains in specific media, incubating at 28–30 °C for 16–48 h under mild stirring. The final inoculum of 4 different starter microorganism for each onion skins source (Table 1) was adjusted in 25 mL starter solution (casein peptone 0.5 g/L; soy peptone 0.5 g/L; NaCl 2 g/L) to obtain a final value corresponding to 7 log10 CFU/ml for yeasts, and 8 log10 CFU/ml for bacteria and added to the heat-treated feedstock mixture. The fermentation was carried out for ten days at 30 °C under aerobic static conditions.
Microbiological, physical-chemical analyses, enzyme activity assay of fermented onion skins
Microbiological assays
Microbiological analyses of samples were carried out following the procedure described by Ramires et al. (2022) with some modifications. Ground onion skins were incubated 1 g/L (w/v) peptone water (Sigma-Aldrich, Darmstadt, Germany) for 1 h under agitation (150 rpm) at room temperature; an aliquot of liquid fraction of heat treated onion skins and fermented samples were diluted with 1 g/L (w/v) peptone water and analyzed on agar media plates:
-
Plate Count Agar (PCA, Heywood, Lancashire, UK) with 0.05 g/L nystatin (Sigma-Aldrich, Darmstadt, Germany) and incubated for 48–72 h at 30 °C was used for total bacterial count;
-
Violet Red Bile Glucose Agar (VRBGA, LABM, Heywood, Lancashire, UK) incubated at 37 °C for 18–24 h, for Enterobacteriaceae;
-
Violet Red Bile Agar (VRBA, LABM, Heywood, Lancashire, UK) incubated at 37 °C for 24–48 h, for coli–aerogenes bacteria; Baird Parker Agar Base (BP, LABM, Heywood, Lancashire, UK) incubated at 37 °C for 24–48 h, for Staphylococci;
-
Bacillus ChromoSelect Agar (BCSA, Sigma-Aldrich, Darmstadt, Germany) with Polymyxin B supplement (Sigma-Aldrich, Darmstadt, Germany) incubated at 30 °C for 24–48 h, for Bacillus spp.;
-
Pseudomonas agar (LABM, Heywood, Lancashire, UK) added with CFC supplement (LABM, Heywood, Lancashire, UK) incubated at 35 °C for 24–48 h, for Pseudomonas spp.;
-
Sulphite-Polymyxin-Sulphadiazine Agar (SPS, Biolife Italiana srl, Milano, Italy) incubated at 35–37 °C for 18–48 h under anaerobic conditions, for the detection of Clostridium spp.;
-
Acetobacter spp. were grown for 48–72 h at 28 °C on modified GYC medium (mGYC) containing 50 g/L glucose (VWR Chemicals, Leuven, Belgium), 10 g/L yeast extract (Merck KGaA, Darmstadt, Germany), 2 g/L CaCO3 (Sigma-Aldrich, Darmstadt, Germany) (Hommel 2014) and 0.05 g/L nystatin (Sigma-Aldrich, Darmstadt, Germany).
-
Yeast total count was determined on Sabouraud dextrose agar medium (LABM, Heywood, Lancashire, UK) added with 0.1 g/L of ampicillin (Sigma-Aldrich, Darmstadt, Germany) and 0.05 g/L of kanamycin (Sigma-Aldrich, Darmstadt, Germany) at 30 °C for 3–4 days.
pH and liquid/solid ratio
Fermented and non-fermented samples were filtered by polyamide filter 355/51 (Saati, Milan Italy) using a vacuum pump in order to separate solid and liquid portions. The solid and liquid fractions for each sample were measured by weighing and the results have been expressed as their ratio. The pH was measured by the pHmeter (Hanna Instruments srl, Ronchi di Villafranca Padovana, Italia).
Enzymatic activity assays
Enzyme activity assays for α-amylase, protease, esterase, lipase, cellulase and endo-xylanase were carried out to evaluate the amount of these enzymes deriving from both microorganisms and plant cells in the liquid portion of the samples. All experiments were conducted in triplicate.
For crude enzyme solution preparation, an aliquot of each fermented and unfermented sample was filtered by polyamide filter 355/51 (Saati, Milan, Italy), and the resulting liquid fraction centrifuged at 15,000 x g for 15 min at 4 °C and the resulting supernatant was used for the assays.
α-Amylase activity assay
The α-amylase assay was performed as described by Ramires et al. (2022). Briefly, the reaction mixture consisted of 50 µL substrate solution (1% potato starch in pH 7 phosphate buffer), 93 µL of phosphate buffer at pH 7 and 72 µL of raw enzyme solution. One Unit (U) of activity of α-amylase is defined as the amount of enzyme required to release one micromole of glucose reducing-sugar equivalents per minute.
Protease activity assay
The method by Walter (1984) and Moyano et al. (1996), modified with the use of casein (0.66% (w/v) in 50 mM Tris-HCl buffer pH 8 as a substrate, as proposed by Sigma-Aldrich Company, was followed for this activity test. Protease enzyme activity was measured as a change in absorbance at 765 nm using the microplate reader Infinite M200 PRO (Tecan, Switzerland). One unit of enzyme activity was expressed as 1 µmol of tyrosine min− 1 mg protein− 1.
Esterase activity assay
The carboxyl ester hydrolase (esterase) activity was determined using a modified spectrometric method (Lopes et al. 2011) following the hydrolysis of p-nitrophenylbutyrate (p-NPB) to p-nitrophenol at 37 °C for 5 min. One unit (U) of esterase activity was defined as the amount of esterase needed to release 1 µmol of p-nitrophenol per minute from p-NPB.
Lipase activity assay
A spectrometric method was also performed for the measurement of lipase activity using p-nitrophenyl palmitate (p-NPP) as a substrate (Park et al. 2007). The assay was performed on 1 mL of crude enzyme solution properly diluted as described by Maiorano et al. (2022). One unit (U) of lipase activity was defined as the amount of lipase required to release 1 µmol of p-nitrophenol from p-NPP in 1 min under the corresponding conditions.
Endo-xylanase activity assay
This assay specifically detects the activity of endo-xylanase and not the activity of the enzyme xylosidase or exo-xylanase. The endo-xylanase activity of the supernatants was assayed using Xylanase Assay kits (XylX6 method) (Megazyme, Bray, Ireland), by following the manufacturer’s instructions as described by Maiorano et al. (2022). The endo-xylanase activity (one unit) was defined as 1 µmol of pNP released from XylX6 per minute.
Cellulase activity assay
The catalytic activity of microbial cellulases was tested using a cellulase assay kit (CellG5, Megazyme, Bray, Ireland), as the method provided by the manufacturer, and as described by Maiorano et al. (2022). The cellulase activity (one unit) was defined as 1 µmol of pNP released from CellG5 per minute.
Phenolics content analyses
Polyphenols extraction
Fermented samples were lyophilized and about 0.50 g of each dried sample were mixed with 20 mL of 60% EtOH (ethanol/water 60:40) containing 0.5% v/v trifluoroacetic acid (TFA). The mixtures were kept under constant stirring (150 rpm) for 30 min at room temperature and the extracts were collected after centrifugation at 4500 x g for 10 min. The extractions were performed twice adding 10 mL of fresh solvent to the pellets and the supernatants were combined for further analysis.
Determination of total phenol content
Total Phenol Content (TPC) in the extracts was determined by the Folin-Ciocalteu colorimetric method as reported by Cicco et al. (2009) with slight modifications. Briefly, 100 µL of each extract was mixed with 500 µL of Folin-Ciocalteu phenol reagent and 500 µL of distilled water. After agitation and 3 min standing 1 mL of 20% sodium carbonate (Na2CO3) and 2.9 mL of distilled water were added. Samples were left in the dark at 40 °C for 20 min and the absorbance was spectrophotometrically measured at 750 nm. The calibration curve was plotted versus concentrations of gallic acid ranging from 25 to 400 µg/mL, used as a standard. The results were expressed as mg of gallic acid equivalents (GAE) per gram of dry sample (DW). Two parallel determinations of each sample were performed and average values were calculated.
HPLC-DAD analysis
Aliquots of hydroalcoholic extracts, filtered on 0.45 μm with CA filters, were analyzed by HPLC-DAD using the Agilent 1260 Infinity Series Chromatograph system, supplied with Agilent Open Lab CDS Chem Station Software (Palo Alto, CA, USA). The instrument was equipped with 1260 HIP Degasser, G1312B binary Pump, G1316A Thermostat and G4212B DAD Detector. For separation, analytical 5 μm Phenomenex Luna C18 (4.6 × 250 mm) column (Phenomenex Torrance, CA, USA) was used. The mobile phase was methanol (solvent A) and acetic acid/water (5:95 v/v) (solvent B) and the gradient profile was: 0–25 min, 15–40% A, 25–30 min, 40% A (isocratic), 30–45 min, 40–63% A, 45–47 min, 63% A (isocratic), 47–52 min, 63–100% A, 52–56 min, 100% A (isocratic), with a constant flow of 1 mL/min. Furthermore, the identification of the the main phenolic compounds was performed comparing the spectra and retention time of the pure available standards, as reported by D’Antuono et al. (2018).
Statistical analysis
All data represent the mean of at least three independent replicates (n = 3). Data are presented as mean values ± standard error of the mean values. For microbiological analyses, t-test assay was applied to compare differences between microbial counts; for L/S ratio and enzymatic activities, Kruskal-Wallis test followed by Dunn’s multiple comparisons test were applied to establish significant differences among means values (p < 0.05). Statistical comparisons were performed using Prism 6 software (La Jolla, CA, USA, www.graphpad.com).
The mean values related to phenolic content were subjected to one-way ANOVA followed by Fisher’s post hoc test. Results analysis was performed using the software STATISTICA 6.0 (StatSoft, Tulsa, OK) and was considered as significantly for p ≤ 0.05. The OriginPro 2016 software (OriginLab, USA) was used for further analyses. Principal component analysis (PCA) was used to compare microbiological, chemical, biochemical parameters associated with the fermented and unfermented samples.
Results and discussion
Microbial screening and selection of best candidates for fermentation experiments
To guide the efforts to assess a simplified method for establishing best microbial strains able to use onion skins as the unique fermentation substrate and possibly improve the following extraction of bioactive compounds (polyphenols) from this by-product, a wide range of bacterial and yeast species were assayed. A total number of 94 different bacterial and of 45 yeast strains belonging to the CNR-ISPA collection were used for the screening on the two feedstocks (red and yellow onion skins).
As the first step toward the selection of functional microorganisms, a simple growth medium, prepared with different percentages of onion skins and a negligible quantity of additional nutrients, was set up to check the growth capabilities of different strains.
Microbial growth was measured by applying an arbitrary scale (as described in the Material and Methods section). A query database was elaborated reporting the growth scores of the entire microbial collection used in this study on the tested feedstocks.
In this study, the obtained growth scores were used to confer a Weighted Mean Growth (WMG) value to each strain. By this score the ability of each microbial strain to grow at increasing feedstock concentrations was pondered and encouraged. The WMG value was used to select the best-performing strains as candidates starters to drive the subsequent step of onion skins submerged fermentation experiments (Table S1 and S2).
The analysis of WMG for bacteria ranged from 0 to 3.8 with an average value of 0.7 for red onion skins, and from 0 to 3.5 with an average value of 0.4 for yellow onion skins. Among bacteria, a number of 37 and 68 strains did not grow on agar plate supplemented with at least the 15% hydrated red and yellow onion skins, respectively, and for this reason they were not considered anymore during the study. Only the Acetobacter tropicalis G25 b strain showed a score near 4 for red and yellow onion skins.
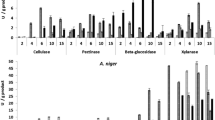
Among Proteobacteria, the average WMG resulted 0.7 and 0.07 for red and yellow onion skins, respectively, whereas for Bacillota (syn. Firmicutes) these values were 0.7 and 1.05 (Table S1). These evidences revealed that tested members of Bacillota phylum were more able to adapt for growth in presence of these two peculiar feedstocks than strains belonging to Proteobacteria and that the two considered Phyla showed a broader range of sensitivity to yellow onion skins than to red ones. The ability to grow in presence of onion skins as main nutrient source varied among all bacteria genera (Fig. 1).
Acetobacter tropicalis strains were the best performers for both the two types of onion skin samples, followed by Serratia liquefaciens, Staphylococcus pasteurii, Levilactobacillus brevis, Lactiplantibacillus plantarum, Leuconostoc mesenteroides. Pediococcus spp., and finally Paenibacillus spp., whereas Sphingomonas and Bacillus simplex strains showed a preferential growth in presence of yellow onion skins.
All yeast strains tested were able to grow on the plates containing yellow or red onion skins homogenates, albeit at different levels (Fig. 2 and Table S2). Good results were obtained for strains belonging to species Aureobasidium pullulans, Candida spp. (C. boidinii, parapsilosis and tropicalis), Debaryomyces hansenii, Metschnikowia pulcherrima, Pichia anomala, Rhodotorula diobovata, Zygosaccharomyces mrakii.
The analysis of WMG for yeasts showed values varying between 1 and 2.8 with an average value corresponding to 1.6 for red onion skins. The WMG values varied 1.7 and 3.5 for yellow onion skins, reporting an average value of 2.1. These evidences revealed that yellow onion skins as unique carbon source affected yeast growth lesser than red ones.
At the end of the screening, it was observed that, except for some bacterial strains, yeasts always showed higher WMG values than bacteria.
The bioprospecting approach carried out on 139 total microbial strains growing on semi-defined microbiological media including different percentages of red or yellow skins allowed to select some strains showing the highest WMG values (Table 1) useful for subsequent steps of the procedure.
Submerged fermentation of onion skins homogenates
Unpasteurized red and yellow onion skins preparations contained initial counts of Enterobacteriaceae (5.7 log10 CFU/mL for red, 5 log10 CFU/mL for yellow onions), Bacillus spp. (4.5 log10 CFU/mL for red, 4.3 log10 CFU/mL for yellow onions), Staphylococci (3.6 log10 CFU/mL), total bacterial count (6 log10 CFU/mL), yeasts and moulds (6 log10 CFU/mL) for both red and yellow onions.
These results are in accordance with previous observations reporting that onions contamination can occur during handling, processing, and storage by the presence of several microorganism such as Bacillus cereus and its spores, lactic acid bacteria, coliforms, Pseudomonas sp., yeast, and moulds (i.e. Aspergillus sp.) (Pezzutti et al. 2005; Savitha et al. 2022).
Therefore, all fermentation tests were subjected to a heat treatment step to reduce possible interferences on growth and activity of the exogenous starter strains. Indeed, after the heat treatment, no spoilage or potential pathogen microbes were detected in red and yellow onion skins preparations, with the exception of Bacillus spp. ranging from 3.6 ± 0.2 and 3.8 ± 0.3 log10 CFU/mL in red and yellow onion skins, respectively and Enterobacteriaceae (3.1 ± 0.2 log10 CFU/mL) only in red onion skins.
A submerged batch fermentation approach was set up for feedstocks treatment. Few studies have already investigated the possible value of red and yellow onions as the sole substrate for bacterial fermentation (Bisakowski et al. 2007; Kimoto-Nira et al. 2020) and none with yeasts. In this study, the onion skins raw material was homogenized and used to prepare samples to be inoculated with microbial selected strains.
Cultures of the selected bacterial and yeast strains were used to inoculate onion skins homogenates at a final concentration of about 8 log10 CFU/mL for bacteria and 7 log10 CFU/mL for yeasts to allow the fermentation process starting rapidly and finishing in a limited period of time, by also outcompeting the residual autochthonous microbiota of onion skins, especially spore forming bacteria.
The load of yeasts, Acetic acid bacteria (AAB) and LAB during fermentation was monitored at the 4th and 10th day. As reported in Table 2, viable and culturable counts of AAB and LAB were about 5–6 log10 CFU/mL at the 4th day fermentation.
As already demonstrated during the screening step on plates, the bacterial strains of Acetobacter tropicalis and Lactiplantibacillus plantarum are expected to live to acid pH present in the onion skins preparations, varying between 3.83 and 4.4 at the initial stage (Table 3). Indeed, the ability of Acetobacter sp. to survive in very harsh environment, such as extreme acid conditions in grape must and wines, has been reported (Francois and Kappock 2007; Bartowsky and Henschke 2008). Also, the promising ability to persist in severe conditions and to modify vegetable matrices characterized by high polyphenols content, such as olive mill wastewaters, was previously demonstrated for the selected strain Acetobacter tropicalis G25b (Ramires et al. 2020).
Moreover, the use of Lactiplantibacillus plantarum strains has been already proven for a wide range of fermented foods and by-products, remarkably vegetables, where difficult environmental constraints to adapt and survive are present (G-Alegria et al., 2004; Muñoz et al. 2017; Tufariello et al. 2015; Zhou et al. 2020).
At the end of the process the bacterial plate counts reduced (to about 3 log10 CFU/mL at 10th day fermentation), this was possible due to the higher concentrations of polyphenols and the limited availability of nutrients in the samples as a result of microbial activities (Table 2). Indeed, the inhibitory effect of different phenolic compounds (such as tannins, terpenes, alkaloids, etc.) partly attributed to their higher concentrations was determined for different LAB (Piekarska-Radzik and Klewicka 2021; García-Ruiz et al. 2012) and Acetobacter (Chen et al. 2022) strains.
Concerning yeasts, they maintained high count values during and at the end of the process (about 6–7 log10 CFU/mL). These results are not surprising, since they are acidophilic organisms and the more adapted to resist low pH values and the high concentrations of phenols of several challenging niches, such as grape must/wine and olive mill wastes (Bleve et al. 2011, 2016; Liu et al. 2015; Ramires et al. 2020). Also yeasts strains belonging to the species and Candida boidinii, Saccharomyces cerevisiae and Metschnikowia pulcherrima are generally adapted to develop in presence of stress conditions and to produce positive effects on the phenolic compounds (i.e. anthocyanins, hydroxycinnamic acids, flavonols, etc.) (Kelanne et al. 2020; Minnaar et al. 2019; Ramires et al. 2020). In addition, the yeast Zygosaccharomyces mrakii strain was isolated and selected from fermented olives and already showed their ability to adapt and survive under difficult environmental constraints (Bleve et al. 2014; Tufariello et al. 2015).
As a preliminary evidence, the liquid/solid ratio showed a statistically significant increase in treated yellow skins (C. boidinii A5y code 33 and S. cerevisiae code 71, p < 0.05), thus suggesting a potential degrading microbial activity of the plant cell walls of these feedstock (Table 3).
Bio-chemical analyses
Enzymatic assays
Previous studies demonstrated that bacteria and yeasts produce many different types of enzymes during fermentation, e.g., glycoside hydrolase, cellulose- or xylan-degrading enzymes, and esterase, which break down plant cell walls or starch and depolymerize polysaccharides (Hur et al. 2014).
In comparison with the untreated samples (unfermented T0, Tables 3 and 4), in yellow and red onion skins inoculated with the selected microbial strains, specific enzyme profiles were obtained (Tables 4 and 5). The reported enzyme activities are the sum of the two components within each sample, those deriving from the onion skins tissues and those produced by the microorganisms. A significant increase (p < 0.05) of endo-xylanase activities was detected in all fermented red onion skins samples, whereas the yellow ones showed increased α-amylase activities (p < 0.05). Interestingly, the red and yellow onion skins samples inoculated with the same Acetobacter tropicalis G25b strain showed also an increase in lipase and endo-cellulase activities (about 1.4 and 1.6-fold, p < 0.05). The inoculation of Lactiplantibacillus plantarum strains was associated to an increased esterase (1.6–2.4 fold, p < 0.05) and lipase (1.3–1.6 fold, p < 0.05) activities in both fermentations, while in presence of L. plantarum TB 11–32 and of L. plantarum C 180 − 34 strains increased protease and endo-cellulase (1.7 fold, p < 0.05) activities were registered, respectively. Finally, a significant improvement in esterase activities was measured in presence of the yeast Zygosaccharomyces mrakii CL 30 − 29 strain in red (1.6 fold, p < 0.05) and of the Saccharomyces cerevisiae En SC strain in yellow onion (2.6 fold, p < 0.05) skins.
Higher enzyme activities in fermented onion skins can be considered as a good indicator of microbial starters active metabolism performed during fermentation of a source different from their original habitat. Indeed, the enzymatic profile expressed in terms of lipase, protease, α-amylase, esterase, endo-cellulase and endo-xylanase activities have been studied in other fermented food products inoculated with selected bacterial and yeast starters (Jolly et al. 2014; Maiorano et al. 2022; Ramires et al. 2022). In addition, these activities were already correlated with the release of bioactive compounds, mainly polyphenols, and with the antioxidant activity, in various plant-based foods (Hur et al. 2014).
Polyphenols characterization of fermented onion skins
The qualitative and quantitative characterization of the bioactive compounds mainly polyphenols, identified after microbial pretreatments, was performed by following Folin-Ciocalteu Assay and HPLC-DAD analysis.
After 10 days of fermentation, a significant increase of total phenol content was observed in the red onion skins fermented samples compared to the unfermented control (T0). In particular, the highest content of total polyphenols was present in the sample inoculated with L. plantarum TB-11-32 strain (Lp_30), reaching 90.96 ± 3.74 mg GAE/g DW (p < 0.05) (Table 6), although no significant differences were observed among all the fermented samples.
Regarding the yellow onion skins, the fermentation process leaded to an increase of the total polyphenol content in all considered samples and in a range moving from 26.9 to 34.5%, expecially for the sample inoculated with S. cerevisiae En SC strain (Sc_71), that showed the highest content (34.24 ± 0.36 mg GAE/g DW, p < 0.05) (Table 6). As was reported for rye bran, brown algae, mulberry, blueberry, yeasts have demonstrated a great potential for the bioactive compounds release; other study demonstrated an increase of total phenolics contents after fermentation by Saccharomyces spp. strains (Hur et al. 2014).
Both the red and yellow onion skins extracts have been also characterized for their polyphenol profile by HPLC-DAD analysis.
In the red onion skins, the main polyphenols identified in all the fermented samples belonged to the classes of flavonoids, phenolic acids and anthocyanins (Fig. 3). In particular, the quercetin aglycone was the most abundant, followed by the glycosidic forms of quercetin that for their low relative abundance have been quantified as quercetin glycosides (Fig. 3).
Polyphenols characterization and quantification (mg/g DW) of red onion skins extracts by HPLC-DAD analysis. 14: Acetobacter tropicalis G25b; 30: Lactiplantibacillus plantarum TB 11–32; 45: Metschnikowia pulcherrima 2047; 57: Zygosaccharomyces mrakii CL 30 − 29; T0: unfermented onion skins as control. Data are represented as mean ± standard deviation (n = 3). Different letters above the columns indicate significant difference (p < 0.05) among the means of each polyphenol class as determined by one-way ANOVA followed by Fisher’s post hoc test
Several research investigation reported the quercetin glycosides conjugated have showed lower antioxidant activity than that quercetin aglycone (Manach et al. 1998). The bioavailability is also affected by the sugary moiety, showing a lower absorption of glycosydes forms respect the aglycone ones (Yang et al. 2012). Hence, the microbial conversion of quercetin glucoside into quercetin aglycone could be a promising strategy to enhance the bioactivity of onion polyphenols. Other identified polyphenols were the dihydroflavonol taxifolin and protocatechuic acid present both for almost 10% of total polyphenols identified, and already identified in onion cultivars (Cattivelli et al. 2023). Recent scientific evidences have reported that the taxifolin rich food can influence brain function also reducing stress (Shinozaki et al. 2023). The anthocyanins (cyanidine derivatives) were about 5% of the total polyphenols identified and they did not undergo variations probably due to the acidic environment of fermented products. Notewhortly to underline the significant differences among the identified polyphenols in the fermented samples in comparison with the unfermented control (T0). In particular, in the samples 57 (fermented by yeast Z. mrakii CL 30 − 29) the quercetin aglycone increase of about 25% (significant difference p < 0.05) with a simultaneous reduction of glycosides quercetins, probably due to the selective deglycosylation reaction occurred on the glycosylic moiety of quercetin glycoside, as reported for S. cerevisiae in onion, by Chung et al. (2011) (Fig. 3).
Taxifolin, protocatechuic acid and glycosylated and aglycone quercetins have been identified also in fermented yellow onion skins by HPLC-DAD analysis (Fig. 4). The results showed that the microbial treatment modify significantly the relative abundance of the identified polyphenols (significant difference p < 0.05) compared to the unfermented sample (T0).
Polyphenolic characterization and quantification (mg/g DW) in yellow onion skins extracts by HPLC-DAD analysis. 14: Acetobacter tropicalis G25b; 28: Lactiplantibacillus plantarum C 180 − 34; 33: Candida boidinii A5y; 71: Saccharomyces cerevisiae En SC; T0: unfermented onion skins as control. Data are represented as mean ± standard deviation (n = 3). Different letters above the columns indicate significant difference (p < 0.05) among the means of each polyphenol class as determined by one-way ANOVA followed by Fisher’s post hoc test
In particular, the fermented sample with A. tropicalis G25b (code 14) showed the higher amount of taxifolin and protocatechuic acid (4.54 and 7.82 mg/gDW, respectively) (p < 0.05); instead, the sample fermented with L. plantarum C 180 − 34 (code 28), underlined the greater increase of quercetin aglycone of about 60% (p < 0.05) compared to the unfermented control.
The reported effect, already highlighted in the red onion skins, can be attributed to lactic acid bacteria (LAB) that can survive to the high polyphenols amount also modificating, by action of a wide enzymatic activities and of the pH, the complex phenolics structure increasing the amount of single compound, also enhancing the health promoting effects of fermented food (Khubber et al. 2022; Kimoto-Nira et al. 2020; Tsangalis et al. 2002). In addition, in the same sample, the protocatechuic acid underwent to a strong degradation probably for the specific microbial metabolism of the selected strain L. plantarum C 180 − 34 (Othman et al. 2009).
Principal component analysis
Principal component analysis was used to apply an unsupervised approach for analyzing the existing relationship among the chemical, physical and biochemical datasets produced during the study and the tested samples of unfermented and fermented red and yellow onion skins. A total variance of 73.54% was observed for red onion skins (Fig. 5).
Score scatter plot of PCA model performed on parameters associated with all red onion skin fermented samples. PCA variables were the data obtained from the analysis of values of chemical composition, nutritional traits, enzyme-associated activities. At_14: Acetobacter tropicalis G25b; Lp_30: Lactiplantibacillus plantarum TB 11–32; Mp_45: Metschnikowia pulcherrima 2047; Zm_57: Zygosaccharomyces mrakii CL 30 − 29; T0: unfermented onion skins as control
The two planes made using the first two principal components (PCs) showed clustering of the red onion skins samples into three groups. One group, consisting of samples treated with the bacterial strain L. plantarum TB 11–32 (Lp_30), with the A. tropicalis G25b (At_14), and with the yeast strain M. pulcherrima 2047 (Mp_45), were closely associated to the parameters total phenolic content (TPC), taxifolin and protocatechuic acid, and, lipase, cellulase and protease activities.
The second group consisted of the sample treated with Z. mrakii CL 30 − 29 (Zm_57) located in the opposite portion of the plane (and mainly associated to pH, quercetin aglycone, esterase and xylanase activities). The untreated sample (T0) was located on the negative semi-axis of the first component, discriminating it from all the other treatments.
Concerning yellow onion skins, a total variance of 71.91% was reported (Fig. 6) and also in this case, three groups were identified.
Score scatter plot of PCA model performed on parameters associated with all yellow onion skin fermented samples. PCA variables were the data obtained from the analysis of values of chemical composition, nutritional traits, enzyme-associated activities. At_14: Acetobacter tropicalis G25b; Lp_28: Lactiplantibacillus plantarum C 180 − 34; Cb_33: Candida boidinii A5y; Sc_71: Saccharomyces cerevisiae En SC; T0: unfermented onion skins as control
The samples treated with the inoculated bacterial and yeast strains were clearly separated by the untreated one (T0). The samples treated with yeast strain C. boidinii A5y (Cb_33) and the bacterial strains L. plantarum C 180 − 34 (Lp_28) and A. tropicalis G25b (At_14) grouped together and were characterized by quercetin aglycone, taxifolin, pH, lipase, cellulase and a-amylase activities. The second group represented by S. cerevisiae En SC (Sc_71) treated sample was mainly associated to TPC and esterase activity.
The choice of microorganism to be used in the fermentation process depends on the desired end products and/or final applications of the treated samples. However, this study suggests that mostly the release yields of bioactive compounds (i.e. polyphenols) and the production of microbial enzymes of biotechnological interest, can be improved with a suitable choice of microorganism to re-use the byproduct onion skins. Additionally, the preliminary evidences can open the opportunity to further study the possible correlation of microbial hydrolytic enzyme activities (such as α-amylase, cellulase, etc.) to the TPC and then in the release of phenolic, as already demonstrated by Chen et al. (2020) for oat.
Conclusions
In the present work, a new procedure for screening microbial biodiversity able to improve the recovery of bioactive compounds and the production of enzymes of biotechnological interest from the lignocellulosic material onion skins was developed. The fermentation was performed using different bacterial (Lactiplantibacillus and Acetobacter spp.) and yeast (Saccharomyces, Metschnikowia and Candida spp.) strains. The resulting products exhibited different levels of polyphenols and enzymatic activities. In fermented yellow onion skins, the release of total phenolic compounds was remarkable, as it was increased of about 26.9–34.5% in comparison with the untreated sample. The treatments of yellow onion skins with L. plantarum C 180 − 34 and S. cerevisae En SC were the most promising processes in terms of recovery of quercetin aglycone as well as of associated interesting enzymes activities. A weaker effect, but still significant, was observed for red onion skin sample treated with L. plantarum TB 11–32 (> 11% TPC than untreated sample). However, the treatment with the yeast strain Zygosaccharomyces mrakii CL 30 − 29 enhanced the quercetin aglycone content of about 25% of the initial raw material.
By this way, the microbial pre-treatment driven by selected bacterial and yeast strains here developed can be considered as a new method, alternative to the broadly recognized traditional physical and chemical approaches, for enhancing the extraction of polyphenols from onion skins feedstock by a sustainable and low-cost route. The peculiar microbial metabolic activities were also able to significantly enrich the recovery of quercetin aglycone, which is a compound greatly requested for its high antioxidant activity. The here set-up bio-process combination with other technologies and scale-up optimization could further enhance bioactive compounds yield and bioactivity.
Data Availability
All data generated or analysed during this study are included in this published article and its supplementary information file.
References
Barcelos MC, Ramos CL, Kuddus M, Rodriguez-Couto S, Srivastava N, Ramteke PW, Mishra PK, Molina G (2020) Enzymatic potential for the valorization of agro-industrial by-products. Biotechnol Lett 42:1799–1827. https://doi.org/10.1007/s10529-020-02957-3
Bartowsky EJ, Henschke PA (2008) Acetic acid bacteria spoilage of bottled red wine—a review. Int J Food Microbiol 125(1):60–70. https://doi.org/10.1016/j.ijfoodmicro.2007.10.016
Bei Q, Chen G, Lu F, Wu S, Wu Z (2018) Enzymatic action mechanism of phenolic mobilization in oats (Avena sativa L.) during solid-state fermentation with Monascus anka. Food Chem 245:297–304. https://doi.org/10.1016/j.foodchem.2017.10.086
Benítez V, Mollá E, Martín-Cabrejas MA, Aguilera Y, López-Andréu FJ, Cools K, Terry LA, Esteban RM (2011) Characterization of industrial onion wastes (Allium cepa L.): dietary fibre and bioactive compounds. Plant Foods Hum Nutr 66:48–57. https://doi.org/10.1007/s11130-011-0212-x
Benito-Román Ó, Blanco B, Sanz MT, Beltrán S (2020) Subcritical water extraction of phenolic compounds from onion skin wastes (Allium cepa cv. Horcal): effect of temperature and solvent properties. Antioxidants 9(12):1233. https://doi.org/10.3390/antiox9121233
Bisakowski B, Atwal AS, Gardner N, Champagne CP (2007) Effect of lactic acid fermentation of onions (Allium cepa) on the composition of flavonol glucosides. Int J Food Sci 42(7):783–789. https://doi.org/10.1111/j.1365-2621.2006.01268.x
Bleve G, Lezzi C, Chiriatti MA, D’Ostuni I, Tristezza M, Di Venere D, Sergio L, Mita G, Grieco F (2011) Selection of non-conventional yeasts and their use in immobilized form for the bioremediation of olive oil mill wastewaters. Bioresour Technol 102:982–989. https://doi.org/10.1016/j.biortech.2010.09.059
Bleve G, Tufariello M, Durante M, Perbellini E, Ramires FA, Grieco F, Cappello MS, De Domenico S, Mita G, Tasioula-Margari M, Logrieco AF (2014) Physico-chemical and microbiological characterization of spontaneous fermentation of Cellina di Nardò and Leccino table olives. Front Microbiol 5:570. https://doi.org/10.3389/fmicb.2014.00570
Bleve G, Tufariello M, Vetrano C, Mita G, Grieco F (2016) Simultaneous alcoholic and malolactic fermentations by Saccharomyces cerevisiae and Oenococcus oeni cells co-immobilized in alginate beads. Front Microbiol 7:1–17. https://doi.org/10.3389/fmicb.2016.00943
Carmona-Cabello M, Garcia IL, Leiva-Candia D, Dorado MP (2018) Valorization of food waste based on its composition through the concept of biorefinery. Curr Opin Green Sustain Chem 14:67–79. https://doi.org/10.1016/j.cogsc.2018.06.011
Cattivelli A, Nissen L, Casciano F, Tagliazucchi D, Gianotti A (2023) Impact of cooking methods of red-skinned onion on colon metabolic transformation of phenolic compounds and gut microbiota changes. Food Funct. https://doi.org/10.1039/D3FO00085K
Celano R, Docimo T, Piccinelli AL, Gazzerro P, Tucci M, Di Sanzo R, Carabetta S, Campone L, Russo M, Rastrelli L (2021) Onion Peel: turning a Food Waste into a resource. Antioxidants 10:304. https://doi.org/10.3390/antiox10020304
Chen G, Liu Y, Zeng J, Tian X, Bei Q, Wu Z (2020) Enhancing three phenolic fractions of oats (Avena sativa L.) and their antioxidant activities by solid-state fermentation with Monascus anka and Bacillus subtilis. J Cereal Sci 93:102940. https://doi.org/10.1016/j.jcs.2020.102940
Chen C, Wu S, Li Y, Huang Y, Yang X (2022) Effects of different acetic acid bacteria strains on the bioactive compounds, volatile compounds and antioxidant activity of black tea vinegar. LWT 171:114131. https://doi.org/10.1016/j.lwt.2022.114131
Cheng A, Chen X, Jin Q, Wang W, Shi J, Liu Y (2013) Comparison of phenolic content and antioxidant capacity of red and yellow onions. Czech J Food Sci 31(5):501–508. https://doi.org/10.17221/566/2012-CJFS
Chung DM, Chung YC, Maeng PJ, Chun HK (2011) Regioselective deglycosylation of onion quercetin glucosides by Saccharomyces cerevisiae. Biotechnol Lett 33:783–786. https://doi.org/10.1007/s10529-010-0501-8
Cicco N, Lanorte MT, Paraggio M, Viggiano M, Lattanzio V (2009) A reproducible, rapid and inexpensive folin–ciocalteu micro-method in determining phenolics of plant methanol extracts. Microchem J 91:107–110. https://doi.org/10.1016/j.microc.2008.08.011
D’Antuono I, Bruno A, Linsalata V, Minervini F, Garbetta A, Tufariello M, Mita G, Logrieco AF, Bleve G, Cardinali A (2018) Fermented apulian table olives: Effect of selected microbial starters on polyphenols composition, antioxidant activities and bioaccessibility. Food Chem 248:137–145. https://doi.org/10.1016/j.foodchem.2017.12.032
Dey TB, Chakraborty S, Jain KK, Sharma A, Kuhad RC (2016) Antioxidant phenolics and their microbial production by submerged and solid state fermentation process: a review. Trends Food Sci Technol 53:60–74. https://doi.org/10.1016/j.tifs.2016.04.007
FAOSTAT. FAO Statistics Division (2018) Available online: http//www.fao.org/faostat/en/#data (accessed on 17 April 2023)
Francois JA, Kappock TJ (2007) Alanine racemase from the acidophile Acetobacter aceti. Protein Expr Purif 51(1):39–48. https://doi.org/10.1016/j.pep.2006.05.016
G-Alegría E, López I, Ruiz JI, Sáenz J, Fernández E, Zarazaga M, Dizy M, Torres C, Ruiz-Larrea F (2004) High tolerance of wild Lactobacillus plantarum and Oenococcus oeni strains to lyophilisation and stress environmental conditions of acid pH and ethanol. FEMS Microbiol Lett 230(1):53–61. https://doi.org/10.1016/S0378-1097(03)00854-1
Ganguly P, Khan A, Das P, Bhowal A (2021) Cellulose from lignocellulose kitchen waste and its application for energy and environment: bioethanol production and dye removal. Indian Chem Eng 63(2):161–171. https://doi.org/10.1080/00194506.2020.1833765
García-Ruiz A, Cueva C, González-Rompinelli EM, Yuste M, Torres M, Martín-Álvarez PJ, Bartolomé B, Moreno-Arribas MV (2012) Antimicrobial phenolic extracts able to inhibit lactic acid bacteria growth and wine malolactic fermentation. Food Control 28(2):212–219. https://doi.org/10.1016/j.foodcont.2012.05.002
Gontard N, Sonesson U, Birkved M, Majone M, Bolzonella D, Celli A, Angellier-Coussy H, Jang GW, Verniquet A, Broeze J, Schaer B, Batista AP, Sebok A (2018) A research challenge vision regarding management of agricultural waste in a circular bio-based economy. Crit Rev Environ Sci Technol 48(6):614–654. https://doi.org/10.1080/10643389.2018.1471957
Gulsunoglu-Konuskan Z, Kilic-Akyilmaz M (2022) Microbial Bioconversion of Phenolic Compounds in agro-industrial wastes: a review of mechanisms and effective factors. J Agric Food Chem 70(23):6901–6910. https://doi.org/10.1021/acs.jafc.1c06888
Gulsunoglu-Konuskan Z, Karbancioglu-Guler F, Kilic-Akyilmaz M (2021) Development of a bioprocess for production of ellagic acid from chestnut (Castanea sativa Mill.) Waste by fermentation with aspergillus spp. Food Biosci 42:101058. https://doi.org/10.1016/j.fbio.2021.101058
Hommel RK (2014) Acetobacter, in Batt C.A, Tortorello ML. (Academic Press) Encyclopedia of Food Microbiology (2th ed. pp. 3–10) ISBN 9780123847331 https://doi.org/10.1016/B978-0-12-384730-0.00001-X
Hur SJ, Lee SY, Kim YC, Choi I, Kim GB (2014) Effect of fermentation on the antioxidant activity in plant-based foods. Food Chem 160:346–356. https://doi.org/10.1016/j.foodchem.2014.03.112
Jolly NP, Varela C, Pretorius IS (2014) Not your ordinary yeast: non-saccharomyces yeasts in wine production uncovered. FEMS Yeast Res 14(2):215–237. https://doi.org/10.1111/1567-1364.12111
Kelanne N, Yang B, Liljenbäck L, Laaksonen O (2020) Phenolic compound profiles in alcoholic black currant beverages produced by fermentation with Saccharomyces and non-Saccharomyces yeasts. J Agric Food Chem 68(37):10128–10141. https://doi.org/10.1021/acs.jafc.0c03354
Khubber S, Marti-Quijal FJ, Tomasevic I, Remize F, Barba FJ (2022) Lactic acid fermentation as a useful strategy to recover antimicrobial and antioxidant compounds from food and by-products. Curr Opin Food 43:189–198. https://doi.org/10.1016/j.cofs.2021.11.013
Kim HM, Choi IS, Lee S, Yang JE, Jeong SG, Park JH, Hwang IM, Chun HH, Wi SG, Kim JC, Park HW (2019) Biorefining process of carbohydrate feedstock (agricultural onion waste) to acetic acid. ACS omega 4(27):22438–22444. https://doi.org/10.1021/acsomega.9b03093
Kimoto-Nira H, Ohashi Y, Amamiya M, Moriya N, Ohmori H, Sekiyama Y (2020) Fermentation of onion (Allium cepa L.) peel by lactic acid bacteria for production of functional food. J Food Meas Charact 14:142–149. https://doi.org/10.1007/s11694-019-00276-4
Liu X, Jia B, Sun X, Ai J, Wang L, Wang C, Zhao F, Zhan J, Huang W (2015) Effect of initial pH on growth characteristics and fermentation properties of Saccharomyces cerevisiae. J Food Sci 80(4):M800–M808. https://doi.org/10.1111/1750-3841.12813
Lopes DB, Fraga LP, Fleuri LF, Macedo GA (2011) Lipase and esterase: to what extent can this classification be applied accurately? Food Sci Technol 31:603–613. https://doi.org/10.1590/S0101-20612011000300009
Maiorano G, Ramires FA, Durante M, Palamà IE, Blando F, De Rinaldis G, Perbellini E, Patruno V, Gadaleta Caldarola C, Vitucci S, Mita G, Bleve G (2022) The controlled Semi-Solid Fermentation of Seaweeds as a strategy for their stabilization and new food applications. Foods 11:2811. https://doi.org/10.3390/foods11182811
Manach C, Morand C, Crespy V, Demigné C, Texier O, Régérat F, Rémésy C (1998) Quercetin is recovered in human plasma as conjugated derivatives which retain antioxidant properties. FEBS Lett 426(3):331–336. https://doi.org/10.1016/S0014-5793(98)00367-6
Minnaar PP, Du Plessis HW, Jolly NP, Van Der Rijst M, Du Toit M (2019) Non-Saccharomyces yeast and lactic acid bacteria in co-inoculated fermentations with two Saccharomyces cerevisiae yeast strains: a strategy to improve the phenolic content of Syrah wine. Food Chem X 4100070. https://doi.org/10.1016/j.fochx.2019.100070
Moyano FJ, Diaz M, Alarcón FJ, Sarasquete MC (1996) Characterization of digestive enzyme activity during larval development of gilthead seabream (Sparus aurata). Fish Physiol Biochem 15:121–130. https://doi.org/10.1007/BF01875591
Munir MT, Kheirkhah H, Baroutian S, Quek SY, Young BR (2018) Subcritical water extraction of bioactive compounds from waste onion skin. J Clean Prod 183:487–494. https://doi.org/10.1016/j.jclepro.2018.02.166
Muñoz R, de las Rivas B, López de Felipe F, Reverón I, Santamaría L, Esteban-Torres M, Curiel JA, Rodríguez H, Landete JM (2017) Biotransformation of Phenolics by Lactobacillus plantarum in Fermented Foods, In Frias, J., Martinez-Villaluenga, C., Peñas E. (Academic Press) Fermented Foods in Health and Disease Prevention (pp. 63–83) ISBN 9780128023099, https://doi.org/10.1016/B978-0-12-802309-9.00004-2
Niu B, Paulson JN, Zheng X, Kolter R (2017) Simplified and representative bacterial community of maize roots. Proc Natl Acad Sci USA 114(12):E2450–E2459. https://doi.org/10.1073/pnas.1616148114
Oren A, Garrity GM (2021) Valid publication of the names of forty-two phyla of prokaryotes. Int J Syst Evol Microbiol 71(10):005056. https://doi.org/10.1099/ijsem.0.005056
Othman NB, Roblain D, Chammen N, Thonart P, Hamdi M (2009) Antioxidant phenolic compounds loss during the fermentation of Chétoui olives. Food Chem 116(3):662–669. https://doi.org/10.1016/j.foodchem.2009.02.084
Park SY, Kim JT, Kang SG, Woo JH, Lee JH, Choi HT, Kim SJ (2007) A new esterase showing similarity to putative dienelactone hydrolase from a strict marine bacterium, Vibrio sp. GMD509. Appl Microbiol Biotechnol 77:107–115. https://doi.org/10.1007/s00253-007-1134-2
Pezzutti A, Marucci PL, Sica MG, Matzkin MR, Croci CA (2005) Gamma-ray sanitization of argentinean dehydrated garlic (Allium sativum L.) and onion (Allium cepa L.) products. Food Res Int 38(7):797–802. https://doi.org/10.1016/j.foodres.2005.03.007
Piekarska-Radzik L, Klewicka E (2021) Mutual influence of polyphenols and Lactobacillus spp. bacteria in food: a review. Eur Food Res Technol 247:9–24. https://doi.org/10.1007/s00217-020-03603-y
Poe NE, Yu D, Jin Q, Ponder MA, Stewart AC, Ogejo JA, Wang H, Huang H (2020) Compositional variability of food wastes and its effects on acetone-butanol-ethanol fermentation. Waste Manage 107:150–158. https://doi.org/10.1016/j.wasman.2020.03.035
Ramires FA, Durante M, Maiorano G, Migoni D, Rampino P, Fanizzi FP, Perrotta C, Mita G, Grieco F, Bleve G (2020) Industrial scale bio-detoxification of raw olive mill wastewaters by the use of selected microbial yeast and bacterial strains to obtain a new source for fertigation. J Environ Manage 265:110574. https://doi.org/10.1016/j.jenvman.2020.110574
Ramires FA, Bleve G, De Domenico S, Leone A (2022) Combination of solid state and submerged fermentation strategies to produce a New Jellyfish-Based food. Foods 11:3974. https://doi.org/10.3390/foods11243974
Ren F, Zhou S (2021) Phenolic Components and Health Beneficial Properties of onions. Agriculture 11:872. https://doi.org/10.3390/agriculture11090872
Roldán E, Sánchez-Moreno C, De Ancos B, Cano MP (2008) Characterisation of onion (Allium cepa L.) by-products as food ingredients with antioxidant and antibrowning properties. Food Chem 108:907–916. https://doi.org/10.1016/j.foodchem.2007.11.058
Sagar NA, Pareek S (2020) Dough rheology, antioxidants, textural, physicochemical characteristics, and sensory quality of pizza base enriched with onion (Allium cepa L.) skin powder. Sci Rep 10(1):18669. https://doi.org/10.1038/s41598-020-75793-0
Sagar NA, Pareek S, Gonzalez-Aguilar GA (2020) Quantification of flavonoids, total phenols and antioxidant properties of onion skin: a comparative study of fifteen indian cultivars. J Food Sci Technol 57:2423–2432. https://doi.org/10.1007/s13197-020-04277-w
Santiago B, Calvo AA, Gullon B, Feijoo G, Moreira MT, Gonzalez-Garcia S (2020) Production of flavonol quercetin and fructooligosaccharides from onion (Allium cepa L.) waste: an environmental life cycle approach. J Chem Eng 392:123772. https://doi.org/10.1016/j.cej.2019.123772
Savitha S, Chakraborty S, Thorat BN (2022) Microbial Contamination and Decontamination of Onion and its products. Appl Food Res 2:100032. https://doi.org/10.1016/j.afres.2021.100032
Sáyago-Ayerdi SG, Venema K, Tabernero M, Sarriá B, Bravo L, Mateos R (2021) Bioconversion of polyphenols and organic acids by gut microbiota of predigested Hibiscus sabdariffa L. calyces and Agave (A. tequilana Weber) fructans assessed in a dynamic in vitro model (TIM-2) of the human colon. Food Res Int 143:110301. https://doi.org/10.1016/j.foodres.2021.110301
Sharma K, Mahato N, Nile SH, Lee ET, Lee YR (2016) Economical and environmentally-friendly approaches for usage of onion (Allium cepa L.) waste. Food Funct 7:3354–3369. https://doi.org/10.1039/C6FO00251J
Shinozaki F, Kamei A, Shimada K, Matsuura H, Shibata T, Ikeuchi M, Yasuda K, Oroguchi T, Kishimoto N, Takashimizu S, Nishizaki Y, Abe K (2023) Ingestion of taxifolin-rich foods affects brain activity, mental fatigue, and the whole blood transcriptome in healthy young adults: a randomized, double-blind, placebo-controlled, crossover study. Food Funct. https://doi.org/10.1039/D2FO03151E
Slama N, Mankai H, Limam F (2021) Streptomyces tunisiensis DSM 42037 mediated bioconversion of ferulic acid released from barley bran. World J Microbiol Biotechnol 37:1–10. https://doi.org/10.1007/s11274-021-03031-4
Tsangalis D, Ashton JF, McGill AEJ, Shah NP (2002) Enzymic transformation of isoflavone phytoestrogens in soymilk by β-glucosidase‐producing bifidobacteria. J Food Sci 67(8):3104–3113. https://doi.org/10.1111/j.1365-2621.2002.tb08866.x
Tufariello M, Durante M, Ramires FA, Grieco F, Tommasi L, Perbellini E, Falco V, Tasioula-Margari M, Logrieco AF, Mita G, Bleve G (2015) New process for production of fermented black table olives using selected autochthonous microbial resources. Front Microbiol 6:1007. https://doi.org/10.3389/fmicb.2015.01007
Walter HE. Proteinases: Methods with hemoglobin, casein and azocoll as substrates in: Methods of Enzymatic Analysis, Vol. 5, Bergmeyer, Bermeyer HU (1984) J
Yang EJ, Kim SI, Park SY, Bang HY, Jeong JH, So JH, Rhee IK, Song KS (2012) Fermentation enhances the in vitro antioxidative effect of onion (Allium cepa) via an increase in quercetin content. Food Chem Toxicol 50(6):2042–2048. https://doi.org/10.1016/j.fct.2012.03.065
Yin L, Zhang Y, Wu H, Wang Z, Dai Y, Zhou J, Liu X, Dong M, Xia X (2020) Improvement of the phenolic content, antioxidant activity, and nutritional quality of tofu fermented with Actinomucor elegans. LWT 133:110087. https://doi.org/10.1016/j.lwt.2020.110087
Zhou Y, Wang R, Zhang Y, Yang Y, Sun X, Zhang Q, Yang N (2020) Biotransformation of phenolics and metabolites and the change in antioxidant activity in kiwifruit induced by Lactobacillus plantarum fermentation. J Sci Food Agric 100(8):3283–3290. https://doi.org/10.1002/jsfa.10272
Acknowledgements
The authors thank Azienda Agricola Veltri srl (CZ, Italy) for supplying red onion skins (cv Cipolla rossa di Tropea). The authors also wish to thank Dr. Antonia Gallo for critically reading the manuscript and helpful discussion.
Funding
Open access funding provided by ISPA - BARI within the CRUI-CARE Agreement. This work was supported by the project “PHENOLEXA - Benign cascade extrative biorefinery for converting agri-food side streams into high-value polyphenolic bioactives and functional fibres for pharma, cosmeceuticals, nutraceuticals and food products” that has received funding from the Bio Based Industries Joint Undertaking under the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 101023225.
This work was also in part supported by Agritech National Research Center and received funding from the European Union Next-GenerationEU (PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR)–MISSIONE 4 COMPONENTE 2, INVESTIMENTO 1.4—D.D. 1032 17/06/2022, CN00000022) and in part by the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.3 - Call for proposals No. 341 of 15 March 2022 of Italian Ministry of University and Research funded by the European Union – NextGenerationEU; Award Number: Project code PE00000003, Concession Decree No. 1550 of 11 October 2022 adopted by the Italian Ministry of University and Research, CUP B83C22004790001, Project title “ON Foods - Research and innovation network on food and nutrition Sustainability, Safety and Security – Working ON Foods”. It has received financial support from “PON Ricerca e Innovazione 2014–2020”, Asse IV “Istruzione e ricerca per il recupero” Azione IV.4 “Dottorati e contratti di ricerca a carattere industriale su tematiche dell’innovazione”, A.Y. 2022-23, XXXVII Cycle, for the PhD project grant of Annamaria Tarantini.
Author information
Authors and Affiliations
Contributions
FAR, IDA, AC, GB: conceived, designed, performed research, analysed data, and wrote and read the paper. ARB, VL, LDA, AT, AG: performed research and analysed data. FB and LP: performed research, read, and approved the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study did not require ethics approval.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ramires, F.A., Bavaro, A.R., D’Antuono, I. et al. Liquid submerged fermentation by selected microbial strains for onion skins valorization and its effects on polyphenols. World J Microbiol Biotechnol 39, 258 (2023). https://doi.org/10.1007/s11274-023-03708-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-023-03708-y