Abstract
Pseudomonas stutzeri A1501 is a model strain used to study associative nitrogen fixation, and it possesses the nitrogen regulatory NtrC protein in the core genome. Nitrogen sources represent one of the important factors affecting the efficiency of biological nitrogen fixation in the natural environment. However, the regulation of NtrC during nitrogen metabolism in P. stutzeri A1501 has not been clarified. In this work, a phenotypic analysis of the ntrC mutant characterized the roles of NtrC in nitrogen metabolism and the oxidative stress response of P. stutzeri A1501. To systematically identify NtrC-controlled gene expression, RNA-seq was performed to further analyse the gene expression differences between the wild-type strain and the ∆ntrC mutant under nitrogen fixation conditions. A total of 1431 genes were found to be significantly altered by ntrC deletion, among which 147 associative genes had NtrC-binding sites, and the pathways for nitrogen fixation regulation, nitrogenous compound acquisition and catabolism and nitrate assimilation were discussed. Furthermore, the oxidative stress-related gene (katB), which was upregulated by ntrC deletion, was suggested to be a potential target gene of NtrC, thus highlighting the importance of NtrC in nitrogenase protection against oxygen damage. Based on these findings, we propose that NtrC is a high-ranking element in the regulatory network of P. stutzeri A1501 that controls a variety of nitrogen metabolic and oxidative stress responsive traits required for adaptation to complex rhizosphere environments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitrogen is one of the most important limiting elements in the environment. Bacteria have evolved many systems to adjust their cell metabolic systems according to the environmental nitrogen supply (Shimizu 2016). NtrC, a regulator protein of nitrogen metabolism, is ubiquitous in bacteria and represents the global regulator of gene expression in response to nitrogen limitation (Jiang and Ninfa 1999, 2009; Schumacher et al. 2013). In bacteria, NtrC and NtrB constitute a two-component regulatory system of nitrogen metabolism, which is mediated by protein phosphorylation signal conduction (Arcondéguy et al. 2001; Ninfa and Jiang 2005). Once phosphorylated, NtrC binds DNA at specific promoters and activates the transcription of target genes (Weiss et al. 1992; Chen and Reitzer 1995).
Homologues of ntrC genes have been found in many nitrogen-fixing bacteria, but the role of NtrC in nitrogen fixation is not essential. NtrC activates the transcription of nifLA in Klebsiella pneumoniae (Merrick 1983; Minchin et al. 1989); however, in Azotobacter vinelandii, Bradyrhizobium japonicum, and Azospirillum brasilense, NtrC is not involved in the expression of nif gene expression but plays a role in other aspects of nitrogen assimilation, such as nitrate utilization and glutamine synthase (GS) activity (Toukdarian and Kennedy 1986; Martin et al. 1988; Liang et al. 1993). In addition, the nitrogen assimilation control gene nac of K. pneumoniae is also regulated by NtrC (Collins et al. 1993). In Rhodobacter capsulatus, NtrC is necessary for urea utilization (Masepohl et al. 2001). The NtrC protein was also found to regulate the biosynthesis of alginate, lipase, and biofilms (Leech et al. 2008; Krzeslak et al. 2008; Kim et al. 2009; Cheng et al. 2018). In both Pseudomonas aeruginosa and Pseudomonas fluorescens, NtrBC and CbrAB form a network to control the C/N balance (Li and Lu 2007; Zhang and Rainey 2008; Sánchez et al. 2017).
Pseudomonas stutzeri A1501was isolated from the rice rhizosphere in China and is a model strain for studying associative nitrogen fixation (Desnoues et al. 2003; Rediers et al. 2003; He et al. 2008). This strain has specific nitrogen metabolic properties, including nitrogen fixation under microaerophilic conditions, denitrification under anaerobic conditions and nitrification under aerobic conditions (Lalucat et al. 2006). The regulation of nitrogen fixation (nif genes) depends on the general nitrogen regulatory system in the core genome (NtrBC and related genes) and the nif-specific regulatory system (NifLA) acquired by horizontal transfer in P. stutzeri A1501 (Yan et al. 2008). In addition, because the associated nitrogen-fixing bacteria and root system could not form root nodules and other special tissue structures, nif gene expression is also greatly affected by environmental factors. It is conceivable that the protein NtrC may participate in the expression of the regulator NifA in P. stutzeri (Xie et al. 2006). However, the role and mechanism of NtrC in the nitrogen metabolism of P. stutzeri A1501 have not been clarified clearly. In this study, we provided evidence that NtrC is required for the regulation of nitrogen metabolism and environmental adaption of P. stutzeri A1501, including nitrogen compound utilization, nitrate assimilation, denitrification and nitrogen fixation, as well as the oxidative stress response. Furthermore, we utilized RNA-seq to compare the global expression profile of the ntrC deletion mutant to the wild type under nitrogen fixation conditions and identified some potential targets regulated by NtrC, such as genes involved in nitrogen fixation, assimilatory nitrate reduction, urea metabolism, electron transport and ammonium transport, demonstrating that NtrC is a global regulator controlling the nitrogen availability of P. stutzeri A1501 under nitrogen fixation conditions. Given the global regulatory role of NtrC in nitrogen-fixing bacteria, A1501 NtrC will be a promising element for studying the synthetic biology of nitrogen fixation systems and enhancing the nitrogen-fixing efficiency of root-associated diazotrophs.
Materials and methods
Bacterial strains and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were cultivated at 30 °C (Pseudomonas stutzeri) or 37 °C (Escherichia coli) in Luria-Bertani (LB) or mineral lactate medium (medium K). The mineral lactate medium (g L−1) contained KH2PO4 (0.4), K2HPO4 (0.1), NaCl (0.1), MgSO4·7H2O (0.2), MnSO4·H2O (0.01), Fe2(SO4)3·H2O (0.01), Na2MoO4·H2O (0.01), C3H5NaO3 (6 mL), and (NH4)2SO4 (0.4). When required, nitrogen sources were supplemented at the following final concentrations: 10 mmol L−1 (NH4)2SO4, 10 mmol L−1 serine, 10 mmol L−1 urea or 10 mmol l−1 KNO3. Cultivation of the strain under anaerobic conditions was carried out using LB medium supplemented with 10 mM KNO3 as a terminal electron acceptor of denitrification. The cultivation vessel, which was 60 mL in volume and contained 20 mL of medium, was sealed with butyl rubber, and the remaining oxygen was removed by flushing the tubes with argon for 10 min. The cultivation vessels were inoculated in triplicate with equal amounts of A1501/A1511 cells and shaken at 200 rpm at 30 °C. At regular time intervals, the OD600 of the 200 µL samples was analysed. Antibiotics were used at the following concentrations: 100 µg mL−1 ampicillin (Amp); 50 µg mL−1 kanamycin (km); 10 µg mL−1 tetracycline (Tc); and 40 µg mL−1 chloramphenicol (Cm).
Construction of the ntrC deletion mutant and complementation plasmids
For ntrC gene replacement, a sacB-based strategy was employed (Schäfer et al. 1994). To construct the ntrC-null mutant (A1511), amplification of a 772 bp DNA fragment located upstream of ntrC was performed using the primer set upF/upR, and amplification of an 806 bp DNA fragment located downstream of ntrC was performed using the set downF/downR (Table S1). Restriction enzyme sites (Bam HI and Hin dIII) incorporated into the oligonucleotide primers to facilitate vector construction are underlined in the oligonucleotide sequences shown in Table S1. An 882 bp DNA fragment containing the Cm resistance cassette was amplified from the plasmid pKatCAT5 by PCR using the primers CmF and CmR. The three amplicons were fused into a 2.46 kb fragment, in which the Cm gene was located between the other two amplicons by overlap extension PCR according to the PCR-based fusion strategy (Shevchuk et al. 2004). The fusion PCR product was then cloned into the multiple cloning sites of the pMD18-T vector (TaKaRa, Japan). The resulting plasmid DNA was double digested with Bam HI/Hin dIII and then cloned into the Bam HI/Hin dIII sites of pK18mobsacB (Schäfer et al. 1994). The resulting plasmid, pK18/delntrC, was mobilized from E. coli into P. stutzeri A1501 by conjugation using pRK2013 (Figurski and Helinski 1979) as the helper plasmid. After mating, the cells were spread on LB plates containing 50 µg/mL Km and 40 µg/mL Cm to screen for clones in which pK18/delntrC had integrated into the A1501 genome via a single recombination event. Another recombination event was then induced to replace ntrC with cat and to remove the Kmr and sacB genes from the genome. A colony of a single recombinant was then grown in nonselective LB medium at 30 °C. Cultures were diluted and spread onto LB agar supplemented with 10% (wt/vol) sucrose and 40 µg/mL Cm. The ntrC mutant strain was selected for the kanamycin-sensitive and sacB-negative colonies. Correct recombination was checked using the primers testF and testR, followed by nucleotide sequencing of the amplicon obtained. The resulting ntrC deletion mutant, A1511, was used for further study.
DNA fragments containing WT genes for ntrC or cbrB with their promoter and terminator regions were amplified by PCR to construct complementation plasmids. Two complementation DNA fragments containing ntrC or cbrB were doubly digested with Hin dIII/Bam HI and then ligated into the broad host plasmid pLAFR3 (Staskawicz et al. 1987). The resulting two complemented plasmids, pLAntrC and pLAcbrB, were used for further studies.
RNA isolation for qRT–PCR
Total RNA was isolated from bacteria cultured under the described conditions using the SV total RNA isolation system (Promega, Madison, WI) according to the manufacturer’s instructions. Total RNA was quantified using microspectrophotometry (NanoDrop Technologies, Inc.). RNA integrity was measured using an Agilent 2100 Bioanalyser (Agilent Technologies, Inc.). RNA samples with RNA integrity numbers (RINs) above 7.0 and threshold cycle (CT) values above 32 were used for qRT–PCR.
Quantitative real-time PCR
The expression levels of selected genes were determined by qRT–PCR with Power SYBR green PCR master mix using an ABI Prism 7500 sequence detection system (Applied Biosystems, USA) according to the manufacturer’s instructions. Primers were designed based on sequences of selected genes, which were imported into OligoPerfect (Invitrogen, USA), a primer design software program designed to generate primer pairs suitable for real-time PCR. Primers used for qRT–PCR are listed in Table S1. All qRT–PCRs were performed in triplicate using a 25-ml mixture containing cDNA (5 ml of a one-fifth dilution), 1× brilliant SYBR green quantitative PCR master mixture (Stratagene, USA), and approximately 5 pmol of each primer. Amplification and detection of specific products were performed using the following procedure: 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s, 55 °C for 1 min, and 72 °C for 30 s and then a dissociation curve analysis. The 16 S rRNA gene was used as the endogenous reference control, and relative gene expression was determined using the 2−ΔΔCT relative quantification method. To obtain a standard curve for real-time PCR (RT–PCR), PCR was performed with each primer set by using calibrated amounts of chromosomal DNA, and these assays were performed at the same time as qRT–PCR.
Nitrogenase activity assays
Nitrogenase activity was determined according to the previously described derepression protocol (Desnoues et al. 2003). Bacterial suspensions were incubated at an OD600 of 0.1 in N-free minimal lactate medium (0.5% oxygen and 10% acetylene) at 30 °C. Protein concentrations were determined using a standard protein assay (Bio–Rad, Hercules, CA) with bovine serum albumin as a standard. The specific activity of nitrogenase was expressed as nmol ethylene per hour per milligram of protein. Each experiment was repeated at least three times.
RNA-seq
Strains A1501 and A1511 were cultured for 5 h under nitrogen fixation conditions. RNA was extracted using TRIzol LS reagent (Invitrogen, USA) following the manufacturer’s instructions. Host-cell RNA was depleted using a MICROBEnrich kit (Ambion, USA), and bacterial 23 and 16 S rRNAs were subsequently depleted with a MICROBExpress bacterial mRNA enrichment kit (Ambion, USA). Total RNA-seq libraries were then constructed and sequenced using an Illumina HiSeq 2500 instrument and the paired-end method by Tianjin Biochip Corporation (Tianjin, China). The raw tag sequence data were analysed for gene annotation, genome annotation, and functional annotation. The quality of all steps was controlled in accordance with the recommendations of Illumina.
Transcriptome data analysis
To analyse the gene expression variation of different samples, the fragments per kb of CDS per million mapped reads (FPKM) value was used to normalize the data and represent the overall gene expression. The differentially expressed genes between the two samples were selected according to their significance based on chi-square tests (P < 0.05, with Bonferroni correction) and at least 2-fold differences. Each transcriptome experiment was repeated independently three times (biological replicates).
Bioinformatics analysis
For the phylogenetic analysis, the amino acid sequences of NtrC proteins from different organisms were obtained from the NCBI. Multiple sequence alignments of full-length proteins were performed using Clustal X (Crooks et al. 2004). The pairwise deletion option was used to circumvent the gaps and missing data. We used the neighbour-joining tree generated by the MEGA (Molecular Evolutionary Genetics Analysis) program with 1000 replicates of bootstrap analysis (Datsenko and Wanner 2000).
For the WebLogo analysis, MEME (http://meme.sdsc.edu/) (Bailey and Elkan 1994) and BioProspector (http://robotics.stanford.edu/~xsliu/BioProspector/) (Liu et al. 2001) were used to perform a sequence analysis of the upstream regions of significantly changed genes. The sequence logo was created with WebLogo (http://weblogo.berkeley.edu/) (Crooks et al. 2004).
Phenotype microarray (PM) analysis
The growth phenotype of the ntrC mutant was assessed using 96-well microtiter PM3 plates (Biolog, Hayward CA), with each well containing the defined medium with a unique nitrogen (PM3) compound plus an indicator dye for cell respiration. Excluding a carbon-free well (negative control) for each plate, the PM3 assay assessed the ability of a bacterium to utilize 95 nitrogen compounds as the sole nitrogen source (succinate is the carbon source). Experiments were performed following the manufacturer’s instructions. A total of 100 mL of this cell suspension inoculated into the Biolog inoculating fluid was transferred into each well and incubated at 30 °C for 24 h. The colour intensity was measured every 15 min using the OmniLog reader.
Abiotic stress-resistance assays
Wild-type A1501, ntrC mutant A1511 and complemented strain A1512 were grown in LB medium at 30 °C to an OD600 of 0.6 and then transferred into fresh LB medium in the presence or absence of 0.5 mM CHP. At the time indicated (oxidative stress, 10 min), 10-fold serial dilutions were performed, and 8 µL of each dilution was spotted onto solid LB plates. These plates were incubated at 30 °C for 24 h before colony growth was observed and enumerated.
Results
Growth analysis of a P. stutzeri A1501 ntrC mutant and a complemented derivative under different nitrogen sources
The genomic sequence analysis showed that P. stutzeri A1501 contains a single copy of a putative ntrC-like gene (PST0349), which has an open reading frame (ORF) of 1436 bp (Yan et al. 2008). In other bacteria, the ntrC gene is located in an operon downstream of the gene coding for its potential sensor kinase NtrB and the gene glnA, which encodes a glutamine synthetase (Liu et al. 2017). Similar to other bacteria, the ntrC-like gene in P. stutzeri A1501 is located downstream of NtrB; however, it is distant from the glnA gene. P. stutzeri has only one gene that codes for a PII homologue, whereas enterobacteria have two paralogues that code for GlnK and GlnB (van Heeswijk et al. 1996). Further phylogenetic analysis indicated that the product of ntrC was highly conserved in Pseudomonas species, and compared with the nitrogen-fixing bacteria, the deduced amino acid sequence of NtrC of A1501 showed the highest similarity (86% identity) to the A. vinelandii DJ NtrC protein; however, it only had 45% similarity to the homologous protein of A. brasilense (Fig. S1).
In bacteria, NtrC was verified as the regulatory player in nitrogen metabolism (Yeom et al. 2010; Kukolj et al. 2020). To study the role of NtrC in A1501, a mutant strain carrying a deletion of the ntrC gene, which was designated A1511, and the functional complement strain, which was designated A1512, were constructed (see methods). While the ntrC mutant grew at a similar rate and to the same final optical density as the wild type in minimal medium containing ammonium sulfate or serine as the sole nitrogen source and sodium lactate as the carbon source, the mutant lost the utilization capacity for nitrate and urea. The complemented ntrC mutant (A1512) was able to reach a final optical density (OD600) similar to that of the wild type (Fig. 1). Our qRT–PCR results showed that when mutant A1511 cells were grown in minimal medium containing nitrate or urea as the sole nitrogen source, the transcription of nitrate assimilatory genes (nasB, nasC and nasG) was significantly reduced compared to that of the wild-type strain. The transcriptional levels of the urease accessory protein-encoding gene ureE and urease-encoding gene ureC were also strongly repressed in the ntrC mutant (Fig. 2). Meanwhile, the NtrC-putative binding site was found in the promoter region of nasB or ureE by bioinformatics analysis (Table S2), suggesting that NtrC might positively regulate nitrate assimilation and urea metabolism of A1501 in a direct manner.
Growth of the wild-type P. stutzeri A1501 (●), ntrC mutant strain A1511 (▲) and functional complement strain A1512 (■) on ammonium sulfate (a), L-serine (b), nitrate (c) or urea (d) as the sole source of nitrogen. Growth was measured in K medium supplemented with ammonium sulfate, L-serine, nitrate or urea (10 mmol·L−1) as the sole nitrogen source. The results are the means and standard errors of three independent cultures
Effect of ntrC deletion on the expression of nitrate assimilation-related genes (a) and urea catabolism-related genes (b). Relative levels of transcripts are presented as the mean values ± standard deviations (SDs) calculated from three sets of independent experiments and normalized to levels in the wild-type strain. The statistical significance of the difference was confirmed by t tests (**P < 0.01)
NtrC affects the metabolic activity of alternative nitrogen sources
To further understand the physiological function of NtrC in nitrogen catabolism, the ability of the wild-type strain and the ntrC mutant to utilize 95 different nitrogen sources was examined using Biolog Phenotype Microarray (PM) assays. The utilization of 24 N sources was found to be affected by ntrC deletion. In 11 cases (nitrate, nitrite, urea, L-cysteine, L-isoleucine, L-leucine, cytosine, thymine, N-acetyl-L-glutamic acid, uracil and uric acid), the ntrC mutant was compromised significantly in terms of substrate utilization. In contrast, with 9 various amines (D-glucosamine, formamide, acetamide, phenylethylamine, ethylamine, N-butylamine, methylamine, putrescine and ammonia), the ntrC mutant showed enhanced metabolic activity compared to the wild-type (Fig. 3). The inability of mutant A1511 to utilize nitrate as a sole nitrogen source suggests that NtrC controls the expression of genes essential for the assimilation of nitrate. Denitrification is one of the most important processes in the bacterial nitrogen cycle. A1501 can use nitrate as an electron acceptor and shows nitrogen fixation activity under anaerobic conditions (Lin and You 1987). To study the role of NtrC in nitrate respiration, we investigated the denitrification ability of the wild-type A1501 and ntrC mutant A1511 under anoxic conditions. The results shown in Fig. S2 confirmed that the ability of mutant A1511 to use nitrate as a terminal electron acceptor was decreased by 70% compared with that of the WT under anoxic conditions. These results suggest that NtrC is essential for nitrogen source utilization under aerobiotic or anoxic conditions.
Role of the ntrC gene in the utilization of nitrogen substrates as determined by Biolog phenotype microarray (PM3) analysis. Signal intensities were measured using an OmniLog detection system and expressed as relative OmniLog units (OLU). Data are the means ± the standard error from at least three independent cultures, assayed in triplicate. The statistical significance of the difference was confirmed by t tests (**P < 0.01; *P < 0.05)
NtrC is required for the positive regulation of nitrogen fixation
NtrC regulates the function of the nitrogenase-specific regulator NifA in K. pneumoniae, although it has no effect on the expression of the nitrogenase complex in A. brasilense and A. vinelandii (Zhang et al. 1997; Wang et al. 2012). To evaluate the role of NtrC in the nitrogenase system of A1501, nitrogen-fixing activity was detected under nitrogen fixation (nitrogen-free and microaerobic) conditions. ntrC deletion resulted in an approximately 90% reduction in nitrogen-fixing activity, and this defect was restored by the introduction of a single copy of ntrC (Fig. 4a). The quantitative real-time PCR (qRT–PCR) results showed that the expression levels of the encoding genes for nitrogenase reductase NifH, nitrogenase specific regulator NifA, nitrogen regulatory PII protein GlnK, ammonium transporter AmtB and glutamine synthetase GlnA were decreased to various extents in the ntrC mutant compared with the wild type, whereas these inductions were fully or partially restored to wild-type levels by the complementation plasmid with a wild-type ntrC gene (Fig. 4b). Furthermore, the conserved putative NtrC-binding site sequence was found in the promoter region of nifA, glnK or glnA, suggesting that their expression might be transcriptionally activated by NtrC and that NtrC positively regulated the nitrogen fixation of P. stutzeri A1501 (Table S2).
NtrC is required for the positive regulation of nitrogen fixation. a Nitrogenase activity in the wild-type A1501 (circles), ∆ntrC A1511 (triangles) and the complemented strains A1512 (squares), A1513 (diamond). b Effect of ntrC deletion on the expression of nif genes and their regulators. Relative levels of transcripts are presented as the mean values ± standard deviations (SDs) calculated from three sets of independent experiments and normalized to levels in the wild-type strain
The two-component systems CbrAB and NtrBC form a network to control the C/N balance in P. aeruginosa (Li and Lu 2007). We found that the complementary strain A1513 (mutant A1511 containing pLAcbrB) could recover the inhibited nitrogenase activity caused by ntrC deletion (Fig. 4a). The results of quantitative real-time PCR (qRT–PCR) showed that the expression levels of nitrogen fixation-related genes were fully or partially restored to wild-type levels by the complementation plasmid with the cbrB gene (Fig. 4b), and this finding strongly indicated that CbrB and NtrC regulate nitrogen fixation in a cooperative manner.
Genome-wide analysis of the NtrC regulatory network in P. stutzeri A1501 under nitrogen fixation conditions
To further identify genes that respond to nitrogen fixation conditions in an NtrC-dependent manner, a global transcriptional profiling analysis was conducted with wild-type A1501 and the null-ntrC mutant A1511 under nitrogen fixation conditions. Compared to the wild type, the expression levels of a total of 1431 genes exhibited more than a twofold change in the mutant A1511 strain under nitrogen fixation conditions. Among these genes, the transcription of 1253 genes was enhanced, and the expression of 178 genes was repressed in the ntrC mutant (DESeq analysis P < 0.05 and fold change > 2.0 or < 0.5). In particular, among these downregulated genes, the 49 kb expression island containing nif and other associated genes was markedly downregulated by ntrC inactivation, indicating the dominant role of NtrC in the nitrogen fixation regulation of P. stutzeri A1501, and these findings are consistent with the phenotypic and expressional analysis described above, thus indicating the reliability of RNA-Seq.
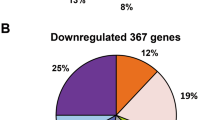
The ntrC mutant resulted in changes in gene expression for several functional categories under nitrogen fixation conditions. These altered genes were further classified according to the COG functional classification system, and the relative occurrence of genes belonging to each category is shown in Fig. 5. Most interestingly, the strong downregulation of genes involved in transport and metabolism enzyme functions (43%) indicated that the deletion of ntrC altered the composition of proteins related to the transport and catabolism of nitrogenous compounds. Furthermore, genes related to energy production and conversion (5%) were upregulated, suggesting that the ntrC mutant might affect the biosynthetic capabilities of the cell under nitrogen fixation conditions.
Overview of expression profiling analysis. a Functional categories of the core subset of upregulated genes (P < 0.05 and fold change > 2) in the ntrC mutant versus wild type under nitrogen fixation conditions. b Functional categories of the core subset of downregulated genes (P < 0.05 and fold change < 0.5) in the ntrC mutant versus wild type under nitrogen fixation conditions. The percentage of genes in each section is depicted
Next, to investigate the potential targets of NtrC involved in nitrogen metabolism, the promoter regions of the 1431 changed genes were analysed. The results showed that 147 NtrC-dependent genes exhibited putative NtrC-binding sites, which contain highly conserved GC and GC elements with an 11-nucleotide spacing, by WebLogo analysis (Fig. S3). Among the 756 top-ranking differentially expressed genes (P < 10−2, fold change > 2.0 or < 0.5), 141 genes were dramatically repressed in the ntrC mutant. The ntrC null mutant resulted in genes involved in nitrogen assimilation and nitrogen fixation, such as glutamine synthetase (glnA, PST0353), PII sensor proteins (glnK, PST0502) and nitrogen fixation regulatory proteins (nifA, PST1313; nifL, PST1314), which showed 0.06-, 0.14-, 0.16- and 0.13-fold reduced transcription, respectively (Table S2). Consistent with the inability of the ntrC mutant to grow with urea or nitrate as the sole nitrogen source, the genes required for urea (ureD-2, ureE, ureF-2, ureG and ureA) and nitrate (nasS, nasT, nasA, nasF, nasD and nasB) transport and utilization displayed strongly downregulated transcription. Additionally, the genes coding for electron transport (rnfABCDGEH) and ammonium transporter (amtB1 and amtB2) were significantly repressed in the ntrC mutant. Since amtB1 and amtB2 are cotranscribed with glnK, which has an NtrC-binding site in the promoter region, we inferred that the transcription of amtB1 and amtB2 is NtrC-dependent. We also found that the transcription of the genes ureE (encoding urease), nasB (encoding nitrite reductase), nasF (encoding nitrate transporter) and rnfA (encoding electron transporter) is NtrC-dependent and has a putative NtrC-binding site in the promoter region, indicating that these genes may be the key genes under NtrC control for nitrate assimilation and urea catabolism. Additionally, the transcription of several genes (PST2280, PST2508 and PST4035) involved in chemotaxis was decreased in the ntrC mutant; in particular, the putative NtrC-binding site was found in the promoter of PST2280, which codes for methyl-accepting chemotaxis receptor proteins (MCPs), and PST2508, which codes for methyl-accepting chemotaxis transducers. Chemotaxis is directed motility used by microbes, which sense chemical cues and relocate towards more favourable environments. Since MCPs are the most common receptors in bacteria, we inferred that NtrC might contribute to the interaction of A1501 with plant hosts. Among the 615 top-ranking genes with dramatically increased transcription, the expression levels of several genes involved in the glycolytic pathway were enhanced significantly in the ntrC mutant, including PST0991, which codes for glucose dehydrogenase; sucD, which codes for succinyl-CoA synthetase; PST3494, which codes for probable glyceraldehyde-3-phosphate dehydrogenase; eda-1, which codes for 4-hydroxy-2-oxoglutarate aldolase; glk-1 and glk-2, which codes for glucokinase; PST3496, which codes for 6-phosphogluconolactonase; PST3497, which codes for glucose-6-phosphate 1-dehydrogenase; and PST3500, which codes for 6-phosphogluconate dehydratase, which showed 3.1-, 2.9-, 6.2-, 9.7-, 3.0-, 5.3-, 4.8-, 6.1- and 11.2-fold increases, respectively (Table S2). Based on these data, we define NtrC as the master nitrogen regulator and infer that it not only activates pathways for nitrogen fixation but also represses carbon catabolism under nitrogen fixation conditions, possibly to prevent excessive carbon and energy flow in the cell.
The ntrC mutant shows altered oxidative stress response
The oxygen concentration is one of the main environmental factors affecting biological nitrogen fixation due to the extreme oxygen sensitivity of nitrogenase. To directly test whether the ntrC mutant displayed altered resistance to oxidative stress, we compared the growth of wild-type strain A1501, the ntrC mutant A1511 and the complementary strain A1512 under oxidative stress conditions by the addition of the oxidizing agent cumene hydroperoxide (CHP). As shown in Fig. S4a, both A1511 and A1512 displayed growth rates similar to that of the wild-type strain in LB medium, indicating that deletion of the ntrC gene had no effect on bacterial survival under normal growth conditions. However, we found that the deletion of ntrC resulted in significantly increased growth in the presence of 0.5 mM CHP, and the complementary strain recovered the growth capacity to the wild-type level under the same treatment (Fig. S4a). Consistent with the observations mentioned above, oxidative stress-related genes were increased to various extents in the ntrC mutant compared with the wild type (Fig. S4b), especially the catalase-encoding gene katB, whose expression was increased 11-fold. Bioinformatic analysis revealed one NtrC-binding site in the katB promoter region, and we inferred that katB is the target gene of NtrC involved in directly regulating optimal oxidative stress resistance.
Discussion
In this study, we used global gene expression and phenotypic analyses to characterize the role of NtrC in the nitrogen metabolism of P. stutzeri A1501 and found that 1431 genes were significantly differentially expressed altered by the ntrC mutant. This large number of differentially expressed genes (33.95% of the genome) shows that a major NtrC-dependent transcriptomic response is initiated by P. stutzeri A1501 under nitrogen fixation conditions. As expected, genes that are known or predicted to be involved in nitrogen metabolism form the majority of the NtrC regulon. In particular, nifA, which codes for the transcriptional activator of all nif operons (Chengtao et al. 2004; Demtröder et al. 2019), showed a 0.16-fold decrease, and glnK, which codes for a PII family protein (Xu et al. 1998; Blauwkamp and Ninfa 2002), showed a 0.14-fold decrease in the ntrC mutant. In P. stutzeri, GlnK is required for both NifA synthesis and activity, particularly by preventing the inhibitory effect of NifL on NifA activity (Xie et al. 2006; He et al. 2008). These data were consistent with the observation that inactivation of NtrC affected nitrogenase activity, suggesting a role in positive regulation of nif genes. Homologues of the ntrC genes have been found in many nitrogen-fixing bacteria, and their roles in nitrogen fixation have been best characterized in K. pneumoniae. In this organism, NtrC plays an important role in the transcription of nifLA regulatory genes, with NifA activating the transcription of other nif operons (Merrick 1983). However, in some diazotrophs, such as A. vinelandii, B. japonicum, and A. brasilense, NtrC is not necessary for nif gene expression. In this study, although NtrC acted as a transcriptional activator of nifA, the mutant was Nif+, and its nitrogen fixing activity was far lower than that of the wild type. NtrC may positively regulate nitrogen fixation, and the CbrB protein may be able to substitute NtrC to maintain nitrogenase activity.
The largest category of genes in the NtrC regulon is the nitrogen scavenging category, which is logical from an evolutionary perspective because the soil-dwelling P. stutzeri A501 encounters various nitrogen sources in the environment and must compete with other soil microbes for nutrients. The genes encoding nitrogen transporters and binding proteins, ammonium transporters, uptake systems for nitrate/nitrite, urea, and amino acids/peptides were all upregulated by NtrC in P. stutzeri A1501 under nitrogen fixation conditions. A similar situation is observed for nitrate/nitrite uptake and assimilation because P. stutzeri A501 contains two nitrate transporters, binding and response proteins (i.e., PST2003, NasA, NasF, NasE and NasD) and nitrite reductase (i.e., NasB and NasC), which are all upregulated by NtrC in P. stutzeri A1501. Assimilatory nitrate reduction to ammonium is a two-step process that includes the reduction of nitrate to nitrite by nitrate reductase followed by the reduction of nitrite to ammonium by nitrite reductase. As confirmed in this study, the assimilatory nitrite reductase NasBC (PST2409 and PST2410) is upregulated by NtrC under nitrogen fixation conditions; however, the nitrate reductase enzyme NasG (PST2411) is not. Therefore, the uptake and assimilation of nitrite, not nitrate, appears to be an important nitrogen stress response in P. stutzeri A501. Our study also identified a NtrC-regulated response regulator, NasT (PST2401). In our previous study, nasT mutant was unchanged the nitrate uptake capacity of P. stutzeri A1501 but could not grow using nitrate as the nitrogen source (unpublished data). However, the precise role of this regulator and nitrate/nitrite respiration in the nitrogen stress response in P. stutzeri A1501 requires further investigation.
The phenotype of the ntrC mutant indicated that NtrC was not only absolutely required for nitrogen metabolism in P. stutzeri but also related to optimal resistance to oxidative stress. The ntrC mutant showed significant upregulation of oxidative stress response genes, especially katB, which is the most pivotal enzyme for the oxidizing agent CHP (Manso et al. 2020). The bioinformatics analysis revealed one NtrC binding site in the promoter region of katB, thus indicating that KatB is a potential target regulated by NtrC. Because oxidative stress is a crucial problem in the survival of nitrogen-fixing bacteria, we inferred that NtrC might regulate oxidative stress resistance via the direct transcriptional activation of katB. In this work, NtrC was shown to be involved in regulating the consumption of some nitrogenous compounds. When it was inactivated and subsequently lost function, the ability to utilize some nitrogenous compounds by P. stutzeri was impaired. These results further confirmed that the ntrC gene was a regulator of the metabolism and assimilation of some nitrogen sources in P. stutzeri. However, not all metabolic pathways for nitrogenous compounds were related to NtrC. For example, the ntrC mutant could grow well with (NH4)2SO4 and some amino acids as the nitrogen source (Glu and Gln). However, the two amino acids were the key signalling molecules in the nitrogen metabolism pathway and switched to nitrogen assimilation in bacteria. Further studies are required to clarify the NtrC-based mechanisms underlying the response of this bacterium to nitrogen signalling and oxidative stress at the cellular and molecular levels.
Taken together, the results of this study provide a framework for understanding the transcriptional changes of numerous key genes related to various nitrogen metabolism processes controlled by NtrC in P. stutzeri A1501 under nitrogen fixation conditions. Chief among these differentially expressed genes are those involved in nitrogen fixation, amino acid catabolism, assimilatory nitrate and ammonium transport. By combining the transcriptome data with bioinformatics analyses, some potential new target genes responsible for electron transport and oxidative stress response regulated by NtrC were discovered, which would help enhance the knowledge of NtrC-based mechanisms underlying both nitrogen metabolism and the environmental adaptation network in P. stutzeri A1501.
References
Arcondéguy T, Jack R, Merrick M (2001) P(II) signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol Mol Biol Rev 65:80–105. https://doi.org/10.1128/MMBR.65.1.80-105.2001
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36
Blauwkamp TA, Ninfa AJ (2002) Physiological role of the GlnK signal transduction protein of Escherichia coli: survival of nitrogen starvation. Mol Microbiol 46:203–214. https://doi.org/10.1046/j.1365-2958.2002.03153.x
Chen P, Reitzer LJ (1995) Active contribution of two domains to cooperative DNA binding of the enhancer-binding protein nitrogen regulator I (NtrC) of Escherichia coli: stimulation by phosphorylation and the binding of ATP. J Bacteriol 177:2490–2496. https://doi.org/10.1128/jb.177.9.2490-2496.1995
Cheng AT, Zamorano-Sánchez D, Teschler JK et al (2018) NtrC adds a new node to the complex regulatory network of biofilm formation and emopenvpsemclose expression in Vibrio cholerae. J Bacteriol. https://doi.org/10.1128/JB.00025-18
Chengtao Y, Guanqiao Y, Shanjiong SSCS, Jiabi Z (2004) Functional difference between Sinorhizobium meliloti NifA and Enterobacter cloacae NifA. Sci China C Life Sci 47:44–51. https://doi.org/10.1360/02yc0268
Collins CM, Gutman DM, Laman H (1993) Identification of a nitrogen-regulated promoter controlling expression of Klebsiella pneumoniae urease genes. Mol Microbiol 8:187–198. https://doi.org/10.1111/j.1365-2958.1993.tb01215.x
Crooks GE, Hon G, Chandonia J-M, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190. https://doi.org/10.1101/gr.849004
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645. https://doi.org/10.1073/pnas.120163297
Demtröder L, Pfänder Y, Schäkermann S et al (2019) NifA is the master regulator of both nitrogenase systems in Rhodobacter capsulatus. Microbiologyopen 8:e921. https://doi.org/10.1002/mbo3.921
Desnoues N, Lin M, Guo X et al (2003) Nitrogen fixation genetics and regulation in a Pseudomonas stutzeri strain associated with rice. Microbiology 149:2251–2262. https://doi.org/10.1099/mic.0.26270-0
Figurski DH, Helinski DR (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA 76:1648–1652. https://doi.org/10.1073/pnas.76.4.1648
He S, Chen M, Xie Z et al (2008) Involvement of GlnK, a PII protein, in control of nitrogen fixation and ammonia assimilation in Pseudomonas stutzeri A1501. Arch Microbiol 190:1–10. https://doi.org/10.1007/s00203-008-0354-x
Jiang P, Ninfa AJ (1999) Regulation of autophosphorylation of Escherichia coli nitrogen regulator II by the PII signal transduction protein. J Bacteriol 181:1906–1911. https://doi.org/10.1128/JB.181.6.1906-1911.1999
Jiang P, Ninfa AJ (2009) Alpha-ketoglutarate controls the ability of the Escherichia coli PII signal transduction protein to regulate the activities of NRII (NtrB) but does not control the binding of PII to NRII. Biochemistry 48:11514–11521. https://doi.org/10.1021/bi901158h
Kim H-S, Park S-J, Lee K-H (2009) Role of NtrC-regulated exopolysaccharides in the biofilm formation and pathogenic interaction of Vibrio vulnificus. Mol Microbiol 74:436–453. https://doi.org/10.1111/j.1365-2958.2009.06875.x
Krzeslak J, Gerritse G, van Merkerk R et al (2008) Lipase expression in Pseudomonas alcaligenes is under the control of a two-component regulatory system. Appl Environ Microbiol 74:1402–1411. https://doi.org/10.1128/AEM.01632-07
Kukolj C, Pedrosa FO, de Souza GA et al (2020) Proteomic and metabolomic analysis of Azospirillum brasilense ntrC mutant under high and low nitrogen conditions. J Proteome Res 19:92–105. https://doi.org/10.1021/acs.jproteome.9b00397
Lalucat J, Bennasar A, Bosch R et al (2006) Biology of Pseudomonas stutzeri. Microbiol Mol Biol Rev 70:510–547. https://doi.org/10.1128/MMBR.00047-05
Leech AJ, Sprinkle A, Wood L et al (2008) The NtrC family regulator AlgB, which controls alginate biosynthesis in mucoid Pseudomonas aeruginosa, binds directly to the algD promoter. J Bacteriol 190:581–589. https://doi.org/10.1128/JB.01307-07
Li W, Lu C-D (2007) Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J Bacteriol 189:5413–5420. https://doi.org/10.1128/JB.00432-07
Liang YY, Arsène F, Elmerich C (1993) Characterization of the ntrBC genes of Azospirillum brasilense Sp7: their involvement in the regulation of nitrogenase synthesis and activity. Mol Gen Genet 240:188–196. https://doi.org/10.1007/BF00277056
Lin M, You CB (1987) Denitrification and nitrogen fixation by Altaligenes faecalis. Acta Agriculturae Nucleatae Sinica 3:3-10
Liu X, Brutlag DL, Liu JS (2001) BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput 2000:127–138
Liu Y, Lardi M, Pedrioli A et al (2017) NtrC-dependent control of exopolysaccharide synthesis and motility in Burkholderia cenocepacia H111. PLoS ONE 12:e0180362. https://doi.org/10.1371/journal.pone.0180362
Manso B, Melero B, Stessl B et al (2020) The response to oxidative stress in Listeria monocytogenes is temperature dependent. Microorganisms. https://doi.org/10.3390/microorganisms8040521
Martin GB, Chapman KA, Chelm BK (1988) Role of the Bradyrhizobium japonicum ntrC gene product in differential regulation of the glutamine synthetase II gene (glnII). J Bacteriol 170:5452–5459. https://doi.org/10.1128/jb.170.12.5452-5459.1988
Masepohl B, Kaiser B, Isakovic N et al (2001) Urea utilization in the phototrophic bacterium Rhodobacter capsulatus is regulated by the transcriptional activator NtrC. J Bacteriol 183:637–643. https://doi.org/10.1128/JB.183.2.637-643.2001
Merrick MJ (1983) Nitrogen control of the nif regulon in Klebsiella pneumoniae: involvement of the ntrA gene and analogies between ntrC and nifA. EMBO J 2:39–44
Minchin SD, Austin S, Dixon RA (1989) Transcriptional activation of the Klebsiella pneumoniae nifLA promoter by NtrC is face-of-the-helix dependent and the activator stabilizes the interaction of sigma 54-RNA polymerase with the promoter. EMBO J 8:3491–3499
Ninfa AJ, Jiang P (2005) PII signal transduction proteins: sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr Opin Microbiol 8:168–173. https://doi.org/10.1016/j.mib.2005.02.011
Rediers H, Bonnecarrère V, Rainey PB et al (2003) Development and application of a dapB-based in vivo expression technology system to study colonization of rice by the endophytic nitrogen-fixing bacterium Pseudomonas stutzeri A15. Appl Environ Microbiol 69:6864–6874. https://doi.org/10.1128/aem.69.11.6864-6874.2003
Sánchez DG, Primo ED, Damiani MT, Lisa AT (2017) Pseudomonas aeruginosa gbdR gene is transcribed from a σ54-dependent promoter under the control of NtrC/CbrB, IHF and BetI. Microbiology 163:1343–1354. https://doi.org/10.1099/mic.0.000502
Schäfer A, Tauch A, Jäger W et al (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. https://doi.org/10.1016/0378-1119(94)90324-7
Schumacher J, Behrends V, Pan Z et al (2013) Nitrogen and carbon status are integrated at the transcriptional level by the nitrogen regulator NtrC in vivo. mBio 4:e00881–e00813. https://doi.org/10.1128/mBio.00881-13
Shevchuk NA, Bryksin AV, Nusinovich YA et al (2004) Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res 32:e19. https://doi.org/10.1093/nar/gnh014
Shimizu K (2016) Metabolic regulation and coordination of the metabolism in bacteria in response to a variety of growth conditions. Adv Biochem Eng Biotechnol 155:1–54. https://doi.org/10.1007/10_2015_320
Staskawicz B, Dahlbeck D, Keen N, Napoli C (1987) Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol 169:5789–5794. https://doi.org/10.1128/jb.169.12.5789-5794.1987
Toukdarian A, Kennedy C (1986) Regulation of nitrogen metabolism in Azotobacter vinelandii: isolation of ntr and glnA genes and construction of ntr mutants. EMBO J 5:399–407
van Heeswijk WC, Hoving S, Molenaar D et al (1996) An alternative PII protein in the regulation of glutamine synthetase in Escherichia coli. Mol Microbiol 21:133–146. https://doi.org/10.1046/j.1365-2958.1996.6281349.x
Wang B, Pierson LS, Rensing C et al (2012) NasT-mediated antitermination plays an essential role in the regulation of the assimilatory nitrate reductase operon in Azotobacter vinelandii. Appl Environ Microbiol 78:6558–6567. https://doi.org/10.1128/AEM.01720-12
Weiss V, Claverie-Martin F, Magasanik B (1992) Phosphorylation of nitrogen regulator I of Escherichia coli induces strong cooperative binding to DNA essential for activation of transcription. Proc Natl Acad Sci USA 89:5088–5092. https://doi.org/10.1073/pnas.89.11.5088
Xie Z, Dou Y, Ping S et al (2006) Interaction between NifL and NifA in the nitrogen-fixing Pseudomonas stutzeri A1501. Microbiology 152:3535–3542. https://doi.org/10.1099/mic.0.29171-0
Xu Y, Cheah E, Carr PD et al (1998) GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition. J Mol Biol 282:149–165. https://doi.org/10.1006/jmbi.1998.1979
Yan Y, Yang J, Dou Y et al (2008) Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc Natl Acad Sci USA 105:7564–7569. https://doi.org/10.1073/pnas.0801093105
Yeom S, Yeom J, Park W (2010) NtrC-sensed nitrogen availability is important for oxidative stress defense in Pseudomonas putida KT2440. J Microbiol 48:153–159. https://doi.org/10.1007/s12275-010-0075-0
Zhang X-X, Rainey PB (2008) Dual involvement of CbrAB and NtrBC in the regulation of histidine utilization in Pseudomonas fluorescens SBW25. Genetics 178:185–195. https://doi.org/10.1534/genetics.107.081984
Zhang Y, Burris RH, Ludden PW, Roberts GP (1997) Regulation of nitrogen fixation in Azospirillum brasilense. FEMS Microbiol Lett 152:195–204. https://doi.org/10.1111/j.1574-6968.1997.tb10428.x
Acknowledgements
This work was supported by the Science and Technology Research Projects of Anhui Province (201904b11020043 and 201904e01020014), the Natural Science Foundation of Anhui Province (1808085MC60), National Key R&D Program of China (No. 2019YFA0904700), the National Natural Science Foundation of China (Nos. 31770067 and 31930004) and the Agricultural Science and Technology Innovation Program of CAAS (No. CAAS-ZDRW202009).
Accession numbers The RNA-seq data described here have been deposited in the NCBI Sequence Read Archive (SRA) database under the accession numbers SAMN16095163 and SAMN16095164.
Author information
Authors and Affiliations
Contributions
Conceptualization, ZY, QL, YY, ML and YZ; Data curation, ZY, QL, XK, YH, SW, FL, YS and SJ; Formal analysis, ZY, QL, YY, XK, YH, SW, FL, YS, SJ, ML, YZ and YZ; Funding acquisition, YY, ML and YZ; Investigation, YZ; Writing—original draft preparation, ZY, YY, YZ and YZ; Writing—review and editing, ZY, YY and YZ; Supervision, ML and YY. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, Z., Li, Q., Yan, Y. et al. Master regulator NtrC controls the utilization of alternative nitrogen sources in Pseudomonas stutzeri A1501. World J Microbiol Biotechnol 37, 177 (2021). https://doi.org/10.1007/s11274-021-03144-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-021-03144-w