Abstract
Trace metal induced stress is an abiotic factor that limits crop yield, having the additional hazard of their accumulation along trophic chain. This fact supposes an emerging problem concerning the health of the population in the case of edible plants such as Cucurbita pepo (zucchini). Most of the plant physiological responses to this adverse situation are regulated by phytohormones, being abscisic acid (ABA) and jasmonic acid (JA) the most important ones, which biosynthesis comprises a key step in this hormone-mediated signaling. In this work, genes involved in ABA and JA biosynthesis have been searched in the zucchini genome, and their expression has been analyzed in leaves of adult plants subjected to Cd- or Ni-induced stress. The results showed the higher sensitivity of zucchini plants to Ni in comparison to Cd, with a higher phenotypic affection and a major decrease of total dry weight. The study of the expression of 12 target genes (5 related to JA biosynthesis and 7 related to ABA biosynthesis), allowed determining a similar genetic response in C. pepo to these metals. The results extend our knowledge of the role of phytohormones on trace metal stress tolerance. Among all the studied genes, the main ones involved in plant responses to trace metal stress were the ABA-related CpAAO3, CpZEP, and CpNCED4, as well as the JA-related CpLOX2, CpOPR3, CpAOS2, and CpJAR1. These results provide relevant information to be used in future breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
At field, plants are constantly exposed to a wide range of both biotic and abiotic conditions, which can constrain their growth and productivity, and eventually may even cause their death. The presence of trace metal elements (TME) is one of these abiotic factors, which consequences befall not only in the limitation of plant growth but also in their incorporation into human and animal bodies through trophic chains. Therefore, their accumulation can be hazardous for animals and humans, considering their high toxicity for a multitude of living beings’ biological processes such as the modification of nucleic acid and protein structures, the oxidative phosphorylation, the osmotic balance, etc. They also cause disorders in the nervous system and damage in organs such as liver, lung, or kidney (Okereafor et al., 2020). In addition, the existence of some TME which also act as plant micronutrients that are required in low quantities (such as Mn, Mo, Cu, Zn, or Ni), can conduct to different plant responses in a dose-dependent way, what reveals the necessity of studying them individually (Chrysargyris et al., 2022).
Among these elements that act as plant micronutrients, two atoms of Ni are required for urease activation, being this enzyme is involved in urea hydrolysis, contributing to its metabolism to ammonia. Additionally, Ni can also activate the enzyme glyoxalase I, involved in the degradation of plant toxic compounds, particularly under stress conditions (Fabiano et al., 2015). However, higher concentration than that required for adequate plant development of this element is commonly found in some soils as a result of human activities, since it is used for the fabrication of batteries, ceramics, pigments, electronics, or coins among others (Begum et al., 2022). On the other hand, the role of Cd as a plant micronutrient has not been reported, and its excess in soils is mainly due to geogenic and anthropogenic sources where mining and metal industry are the main contributors (Kubier et al., 2019). Both elements, Ni and Cd, are considered the most important pollutant TM, and may compete with other essential nutrients in plants, leading to an oxidative stress and decreases in plant growth, transpiration, and photosynthesis rates, among others (Rahi et al., 2022). Plants have developed a variety of physiological and biochemical responses against TME-induced stress, being most of them regulated by the phytohormones abscisic acid (ABA) and jasmonic acid (JA), including (i) the stomatal closure to avoid the excessive transpiration and root water uptake; (ii) the induction of tonoplast transporters to sequestrate the toxic ions inside the vacuoles; (iii) the inactivation of certain membrane transporters to reduce the absorption of TME; (iv) the production of phytochelatins (chelator proteins that are important for heavy metal detoxification); (v) the production of compatible osmolytes such as proline; and (vi) the production of enzymatic and non-enzymatic antioxidant compounds which act as reactive oxygen species (ROS) neutralizers (Saini et al., 2021; Shukla et al., 2017). Specifically, increases of ABA endogenous content have been reported in plants exposed to different TME (Cd, Hg, Cu, Zn, or Pb), suggesting that increased levels of this phytohormone contribute to an improved stress tolerance through the control of the stomatal aperture and transpiration, reducing water consumption and the uptake and intoxication by TME (Bücker-Neto et al., 2017). JA also triggers plant responses to TME, being able to act synergistically with ABA through the modulation of its biosynthesis under abiotic stress conditions (Kim et al., 2021). For instance, exogenous applications of these plant hormones have been reported as mitigators of TME induced stress. Applications of JA in rapeseed (Brassica napus L.) plants subjected to high quantities of Cd resulted in an enhanced tolerance through the maintenance of gas exchange parameters, oxidative status, and pigment content (Ali et al., 2018). Exogenous applications of ABA have been also related with a higher tolerance to high Cd concentrations in several plant species, being reported that ABA contributes to the reduction of Cd ion uptake and transport (Zhao et al., 2023). In this line, the beneficial effect of both phytohormones, JA and ABA, in different plant species under TME stress conditions may indicate that genotypes naturally overproducing these phytohormones may have an enhanced resistance to this harmful situation. The identification of the most important genes in the biosynthesis pathways of these phytohormones could contribute to the selection of target genes for breeding programs focused on the obtention of TME stress tolerant lines. Consequently, these new genotypes would bring the opportunity of maintaining crop yield and productivity without assimilating toxic ions and avoiding the necessity of exogenous applications of these phytohormones, reducing production costs and handling.
Thus, some of the critical genes that codify for key enzymes involved in JA biosynthesis are lipoxigenase 2 (LOX2), that regulates the conversion of linolenic acid to 13-hydroperoxylinolenic acid; allene oxide synthase (AOS), responsible of converting 13-hydroperoxylinolenic acid to 12, epoxy-9, 11, 15-octadecatrienoic acid; oxoohytodienoate-reductase 3 (OPR3), that reduces 12-oxo-cis, cis-10, 15-phytodienoic acid (OPDA) to 3-oxo-2-(2′-pentenyl)-cyclopentane-1-octanoic acid; and jasmonate resistant 1 (JAR1), that regulates the conjugation of jasmonic acid with isoleucine to synthesize jasmonoyl-isoleucine (JA-Ile), reported as one of the main bioactive compounds of JA pathway (Sup. Fig. 1; Wasternack & Song, 2017).
Meanwhile, the key enzymes involved in ABA biosynthesis include zeaxanthin epoxidase (ZEP), that regulates the conversion of zeaxanthin to antheraxanthin and this one to trans-violaxanthin; 9-cis-epoxycarotenoid dioxygenase (NCED), that converts trans-violaxanthin to xanthoxin; abscisic aldehyde oxidase 3 (AAO3), that oxidizes ABA aldehyde to ABA; and cytochrome P450 CYPT707A is considered one of the main regulators of ABA catabolism encoding ABA 8′-hydroxylases (Sup. Fig. 2; Hewage et al., 2020). Regarding ABA biosynthesis and regulation, previous works have concluded that exogenous ABA applications, as well as the activation of ABA biosynthesis-related genes conduct to higher endogenous ABA concentration and enhanced plant resistance to different TME (reviewed in Kumar et al., 2022). Additionally, transgenic Arabidopsis plants overexpressing Malus hupehensis NCED3 accumulate higher ABA amounts and exhibit an enhanced tolerance to Cd, reducing the accumulation of this metal ion (Zhang et al., 2019).
Previous works with Cucurbita pepo L. (zucchini) plants have reported the effects of TME stress in this species, revealing a controversy about if it is a TME hyperaccumulator plant or not, since some works described that this crop accumulates in leaves and roots’ metals such as Ni, Fe, Mn, or Zn at concentrations compatible with its consideration as a hyperaccumulator plant (Galal, 2016), but it seems that trace elements are not translocated to the reproductive organs like flowers (Xun et al., 2017). This fact seems to be in concordance with previous works that remarked the ability of Cd-stressed zucchini plants to avoid the translocation of this toxic ion from roots to some aerial organs. These plants accumulated ABA in roots and fruits although no changes in JA content were detected (Labidi et al., 2021). In addition, the relative expression of genes involved in the synthesis of metallothionines (MT), a metal ion binding protein family, or genes related with the production of antioxidant enzymes responsible of ROS scavenging such as Cu/Zn superoxide dismutase (Cu/ZnSOD), ascorbate peroxidase (APX), or catalase (CAT), were upregulated in C. pepo plants grown under different Cd concentrations (in the range of 50 to 200 μM CdCl2), helping the plant to mitigate the harmful effect of H2O2 (Kar, 2020). However, the biosynthesis pathways of key phytohormones such as ABA and JA under TME stress conditions in this plant species has not been studied yet and supposes a field which can be explored for the selection of candidate genes needed for the design of lines overproducing these two phytohormones, what will confer enhanced TME tolerance.
Soil contamination by TME is an important problem that needs to be studied and evaluated in order to ensure food safety in a world with an increasing population, particularly in some areas polluted by specific TME, consequence of the human activity, as occurs in some soils with high content of Ni (Cetin et al., 2022). In this context, new strategies, including the development of monitoring techniques such as GIS (Cetin et al., 2022) or biomonitors as specific plant species as Cupressus arizonica (Cesur et al., 2022) have been proposed as promising tools. Additionally, the finding and selection of genes related with an enhanced phytohormone biosynthesis may be crucial for future breeding programs conducted to the obtention of plants with an increased capacity to accumulate toxic ions (suitable for bioremediation programs) or a lower metal uptake (favoring the suitability of edible parts of the plant for animal or human feeding). This technique has been extended in the last years in other plant species, reporting a wide range of phytohormone-related genes and proteins which are the key in plant tolerance to heavy metal stress, leading to the obtention of new crop varieties with an enhanced tolerance to this harmful condition (Saini et al., 2021).
The main objective of this work was to elucidate the role of some zucchini key genes on the biosynthesis and regulation of the phytohormones ABA and JA (the main two phytohormones involved in plant responses to abiotic stress) in response to TME-induced stress. As mentioned above, the overexpression of some of these genes can lead to physiological changes, favoring plant survival and the exclusion of toxic ions, and to the best of our knowledge, is an unexplored tool in this crop. For this purpose, we aimed to identify target genes involved in C. pepo JA and ABA biosynthesis which overexpression can contribute to an enhanced tolerance to TME-induced stress. Consequently, in this work adult C. pepo plants were subjected to Ni- or Cd-induced stress for 30 days. Relative expression of 5 target genes related to JA biosynthesis and catabolism and 7 target genes involved in ABA biosynthesis was evaluated. The results indicate the importance of phytohormone regulation in plants subjected to TME-induced stress. Some of the studied genes could be used in future breeding programs to select tolerant varieties to this harmful situation.
2 Material and Methods
2.1 Plant Material, Cultivation, and Experimental Design
The experiments were performed using adult plants of zucchini (Cucurbita pepo L.) and were individually cultured in 0.75 L black plastic pots (12-cm depth and 12-cm diameter), with perlite as inert substrate, as previously described (Labidi et al., 2021). During all the experiments, the plants were kept in a glass greenhouse with a natural photoperiod (13–14 h), with a day/night temperature and humidity of 27/12 ± 5 °C and 60/80 ± 5%, respectively. After seed germination (15 days), plants were watered with 100 mL of modified Hoagland solution (Hoagland & Arnon, 1938) three times per week for 30 days (nutrient solution composition detailed in Sup. Tab. 1). After this period, plants were watered three times per week with 150 mL of the irrigation solution, supplemented with 100, 300, or 500 μM NiCl2 or CdSO4 for 30 days more to induce TME stress by Ni or Cd, respectively, maintaining the greenhouse conditions. An additional group watered with the nutritive solution without adding Ni or Cd sources was included as a non-stressed control and under the same environmental conditions. The concentrations of NiCl2 and CdSO4 were established according to previous works with TME-induced stress in different genotypes, which normally work in the range between 100 and 600 μM (Bankaji, Pérez-Clemente, et al., 2019; Bouslimi et al., 2021; Kouki et al., 2021; Sleimi et al., 2021).
Each one of the seven groups of plants consisted of 10 plants placed randomly in the greenhouse. At the end of the stress period, leaf samples from 5 plants were taken, immediately frozen with liquid nitrogen, ground to fine powder and stored at −80 °C for further analyses. The other five plants from each group were desiccated in a laboratory stove for weighting total biomass production from shoots and roots.
2.2 TME Determination
The quantification of Cd and Ni in leaf and root tissue was performed through atomic absorption spectrometry, as described previously with some modifications (Kouki et al., 2023; Sghaier et al., 2019). Briefly, 50 mg of dry plant tissue sample was digested in teflon bombs with 3 mL of an acidic solution mixture containing HNO3:H2SO4:HClO4 in a 10:1:0.5 v:v:v proportion at 110 °C for 2 h 30 min. After this, samples were taken in 50 mL of 0.5% (v:v) HNO3, filtered, and injected to the atomic absorption spectrometer (PinAAcle 900T, PerkinElmer, Waltham, MA, USA) for determining the content of both elements. The standards of Cd and Ni used for the equipment calibration were prepared from the PerkinElmer solutions of 1000 mg L−1 (PerkinElmer Waltham, MA, USA). The concentrations of Cd and Ni standards in the calibration curve ranged from 0 to 2 mg L−1 for both metals. Final quantification was achieved by interpolation of the values obtained from samples in the calibration curves.
2.3 Gene Search and Primer Design
To study the role of different genes involved in the biosynthesis of JA and ABA, two of the main phytohormones involved in TME stress responses in C. pepo, different genes were selected, including LOX2, AOS, OPR3, and JAR1 as genes involved in JA biosynthesis pathway (Sup. Fig. 1), and ZEP, NCED, AAO3, and CYP707A2 in the case of ABA (Sup. Fig. 2). Gene identification was achieved through the search of genes from the model plant Arabidopsis thaliana in The Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org), obtaining the protein sequence used for running a TBLASTN (protein against nucleotide, translated) in the C. pepo genome database CuGenDB (http://cucurbitgenomics.org/organism/14). The obtained CDS sequences from C. pepo were used for primer design (Montero-Pau et al., 2018). In short, primer design was achieved using Primer3Plus tool (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi; Koressaar & Remm, 2007; Untergasser et al., 2012), checking the absence of homo-dimmers and hetero-dimmer formation with IDT-oligoanalyzer tool (http://eu.idtdna.com/analyzer/applications/oligoanalyzer/), as described in Vives-Peris et al., 2018.
2.4 RNA Isolation, cDNA Synthesis and Gene Expression Analysis
C. pepo total RNA was extracted from frozen plant material, grounded to fine powder, with RNeasy kit (Qiagen, Hilden, Germany) using the procedure provided by the manufacturer. After this, 5 μg of the extracted RNA was treated with DNAse I (Fermentas, Waltham, MA, USA) to degrade the extracted DNA, and 1 μg was converted to cDNA with Primescript RT Reagent Kit (Takara, Shiga, Japan) according to the instructions supplied by the producers. The quality of the extracted RNA and total concentration was estimated with a Nanodrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) after RNA extraction and DNAse treatment (Bankaji, Sleimi, et al., 2019). Finally, gene expression analyses were performed by RT-qPCR using a 48-well ABI StepOne equipment (Applied Biosystems, Foster City, CA, USA), with reactions containing 1 μg of the isolated cDNA, 5 μL of Maxima SYBR Green/ROX qPCR mix (Thermo Scientific), 1 μL of a mix of both primers at 10 μM, and 3 μL of deionized sterile water. The amplification was achieved with a pre-incubation at 95 °C for 10 min and 40 cycles of 10 s of denaturation at 95 °C, 10 s of annealing at 60 °C, and 20 s of extension at 72 °C, as well as a melting curve to check the specificity of the used primers. Actin and tubulin were used as housekeeping genes (Bankaji, Sleimi, et al., 2019).
2.5 Data Processing
For the statistical analysis, data obtained from total dry weight and TME content was processed through one- or two-way analysis of variance (ANOVA), followed by a Tukey post hoc test (p ≤ 0.05) using Infostat 2020 software (Universidad Nacional de Córdoba, Córdoba, Argentina). Meanwhile, the data provided by RT-qPCR analysis were processed with StepOne and StepOnePlus Software v2.3 (Applied Biosystems), and the relative expression was determined with the Relative Expression Software Tool Solver v 2 (REST-MCS; Pfaffl, 2001; Pfaffl et al., 2002). Principal component analysis was performed with SigmaPlot v 14.0 software (Systat Software, Chicago, IL, USA).
3 Results
3.1 Phenotypic Damage
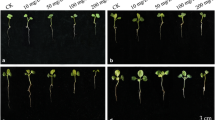
Under greenhouse conditions, the harmful effect of Ni was higher than those observed in Cd-treated plants (Fig. 1). Ni toxicity was evident in C. pepo plants after 30 days of 500 μM Ni application, showing yellow stains in the oldest leaves (Fig. 1a and b), whereas no leaf visual damage was observed in C. pepo plants watered with irrigation solution supplemented with Cd (regardless Cd concentration), which looked similar to control plants (Fig. 1c and d).
Phenotypic damage observed in C. pepo plants after 30 days of TME-induced stress. a General aspect of plants watered with 500 μM Ni; b leaves damaged as a consequence of watering with 500 μM Ni; c general aspect of plants watered with 500 μM Cd; d leaves damaged as a consequence of watering with 500 μM Cd
The highest damage observed in C. pepo plants subjected to Ni-induced stress corresponded with a highest diminution of total shoot and root dry weights in plants treated with this TME (decrease of 84 and 45% in shoots and roots, respectively, in 500 μM Ni-treated plants related to control; Fig. 2a and b) in comparison to those treated with 500 μM Cd, which exhibited a softer dry weight decrease of 72 and 32% in shoots and roots, respectively (Fig 2c and d). Additionally, following these results the threshold concentration which reduced dry weight of zucchini plants was lower for Ni (100 and 300 μM for shoots and roots, respectively) than for Cd (300 and 500 μM for shoots and roots respectively; Fig. 2).
Dry biomass production of C. pepo plants subjected to TME-induced stress at 0 (control), 100, 200, 300, and 500 μM, relative to control. a Shoot dry weight of Ni-stressed plants; b root dry weight of Ni-stressed plants; c: Shoot dry weight of Cd-stressed plants; d: Root dry weight of Cd-stressed plants. Data are the mean of 5 replicates ± standard error. Different letters denote statistically significant differences at p ≤ 0.05
3.2 TME Accumulation
Shoot and root total concentrations of Cd and Ni were boosted as the quantity of these TME was increased in the nutrient solution (Fig. 3). The calibration curves obtained from their quantification are provided in Sup. Fig. 3.
Trace metal ion accumulation of C. pepo plants subjected to TME-induced stress at 0 (control), 100, 200, 300 and 500 μM, relative to control. a: Ni content in leaves from Ni-stressed plants; b: Ni content in roots from Ni-stressed plants; c Cd content in leaves from Cd-stressed plants; d Cd content in roots from Cd-stressed plants. Data are the mean of 5 replicates ± standard error. Different letters denote statistically significant differences at p ≤ 0.05
In shoots of Ni-stressed plants, the content of this element gradually increased from control plants (500 μg g−1 DW) until those treated with 500 μM Ni (1314 μg g−1 DW), what supposes a 2.6-fold increase respect to control (Fig. 3a). Similarly, in roots from plants treated with this element, a 3.0-fold increase was observed comparing the values obtained from roots of control plants with those from plants watered with a 500 μM Ni dose, although no differences in the content of this ion were detected in plants stressed with the lowest Ni dose, 100 μM (Fig. 3b). Additionally, comparing the total amount of Ni between shoots and roots, the content in the former was always slightly higher, independently of the Ni applied dose (Figs. 3a and 3b).
Regarding Cd endogenous content, this element was not detected in control plants, neither in shoots nor in roots (Fig. 3c and d). The different severity of Cd-induced injury was proportionally correlated with its content in leaf tissue. This concentration reached values of 98 and 3424 μg g−1 DW in shoots and roots from plants treated with 500 μM Cd, respectively, being much higher the root content of this ion in comparison to the determined in leaves (Fig. 3c and d).
3.3 Gene Search
A total amount of 12 genes related with JA and ABA biosynthesis were searched in C. pepo genome database. Their accessions and the designed primers are available in Sup. Tab. 2. Regarding JA biosynthesis-related genes, CpLOX2, CpAOS1, CpAOS2, CpOPR3, and CpJAR1 were selected, whereas CpZEP, CpNCED1, CpNCED2, CpNCED3, CpNCED4, CpAAO3, and CpCYP707A2 genes were studied for the ABA biosynthesis pathway.
3.4 Gene Regulation in Plants Subjected to Ni-induced Stress
Differences in the genetic response of JA and ABA biosynthesis related genes after 30 days of exposure to Ni-induced stress were recorded (Fig. 4). In this line, the expression of JA-related genes was altered in plant leaves, being the expression of CpLOX2 importantly repressed in plants watered with Ni in all the studied concentrations, with reductions ranging between 70.5 and 99.9% related to control (Fig. 4a). A similar trend was detected in CpAOS1, with a downregulation ranging between 89.5 and 98.1% in comparison to control (Fig. 4b), although no differences were observed in the expression of the other member of AOS family CpAOS2 (Fig. 4c). Meanwhile, the expression of CpOPR3 was Ni-dose dependent. Although no differences were detected in its expression in leaves of plants watered with 100 μM Ni, it exhibited a 2.6-fold overexpression in plants watered with a 300-μM concentration, and a strong repression with values 79.0% lower than control was registered in plants watered with the highest Ni dose, 500 μM (Fig. 4d). Finally, CpJAR1 was overexpressed in plants from all Ni-treated groups, with expression values ranging between 6.1- and 7.9-fold higher than control (Fig. 4e).
Relative gene expression of JA (a, b, c, d, and e) and ABA (f, g, h, i, j, k, and l) biosynthesis-related genes in zucchini plants subjected to TME stress induced by Ni at 0 (control), 100, 300, and 500 μM, after 30 days. a CpLOX2; b CpAOS1; c CpAOS2; d CpOPR3; e CpJAR1; f CpZEP; g CpNCED1; h CpNCED2; i CpNCED3; j CpNCED4; k CpAAO3; and l CpCYP707A2. The presented data refers to the relative expression compared to control, being the mean of 4 replicates ± standard error. Asterisks refer significant differences compared to control
Regarding ABA biosynthesis-related genes, the gene codifying for the first key enzyme involved in the production of trans-violaxanthin and antheraxanthin from zeaxanthin, CpZEP, was upregulated under all Ni treatments, with values ranging from 47.6- to 248.0-fold higher than those obtained from control plants (Fig. 4f). A similar trend was observed in the expression of other key genes in ABA biosynthesis pathway acting downwards than CpZEP, as CpNCED4 (values from 4.8 to 23.5 times higher than control; Fig. 4j), CpAAO3 (expression between 2.4- and 6.3-fold higher than control; Fig. 4k), or CpCYP707A2 (upregulated between 2.5 and 4.9 times in comparison to control; Fig. 4l) for all Ni treatments. Contrary to the behavior observed in the expression of CpNCED4, other members of this gene family were downregulated under Ni-induced stress independently of the applied concentration, with repressions around 56.7 and 93.6% related to control for CpNCED1 (Fig. 4g), and around 95% for CpNCED3 (Fig. 4i), whereas CpNCED2 was only repressed by Ni when this TME was applied at the highest dose of 500 μM (downregulation of 52.9% respect to control; Fig. 4h).
3.5 Gene Regulation in Plants Subjected to Cd-induced Stress
The expression of genes involved in JA biosynthesis in C. pepo plants subjected to high concentrations of Cd was mostly similar to the observed in plants exposed to Ni (Fig. 5). In this way, JA biosynthesis-related genes CpLOX2 and CpAOS1 in response to Cd-induced stress were repressed independently of the applied concentration, as occurred under Ni stress conditions, with mean downregulations of 69.8 and 88.3% related to control for CpLOX2 (Fig. 5a) and CpAOS1 (Fig. 5b), respectively. Interestingly, contrarily to CpAOS1, CpAOS2 was overexpressed in plants watered with 300 or 500 μM Cd, reaching values 2.6-fold higher than control (Fig. 5c). Downwards in the JA biosynthesis pathway, the gene CpOPR3, responsible to produce the enzyme catalyzing the conversion of OPDA into JA was downregulated, but only at the highest Cd dose of 500 μM, with a repression of 64.3% respect to control (Fig. 5d). Finally, CpJAR1, involved in the catabolism of JA to another bioactive form (JA-Ile) was overexpressed under 100 and 300 μM Cd (4.3- and 6.8-fold higher than control, respectively), although no difference in its expression was recorded after watering plants with 500 μM Cd for 30 days (Fig. 5e).
Relative gene expression of JA (a, b, c, d, and e) and ABA (f, g, h, i, j, k, and l) biosynthesis-related genes in zucchini plants subjected to TME stress induced by Cd at 0 (control), 100, 300, and 500 μM, after 30 days. a CpLOX2; b CpAOS1; c CpAOS2; d CpOPR3; e CpJAR1; f CpZEP; g CpNCED1; h CpNCED2; i CpNCED3; j CpNCED4; k CpAAO3; and l CpCYP707A2. The presented data refers to the relative expression compared to control, being the mean of 4 replicates ± standard error. Asterisks refer significant differences compared to control
As occurred with the regulation of genes involved in JA biosynthesis, those involved in ABA production also exhibited a similar expression pattern under Ni and Cd stress conditions. Examples of this overexpression occurred with CpZEP (overexpression between 3.1 and 14.7-fold higher than control; Fig. 5f), CpNCED1 (its expression decreased between 41.9 and 73.7% regarding control; Fig. 5g), CpNCED3 (repressed in the range of 41.9 and 73.7% compared to control; Fig. 5i), CpAAO3 (overexpression between 2.0 and 13.5 related to control; Fig. 5k), and CpCYP707A2, exhibiting expression values 13.5 and 3.4 times higher than control, but only in plants treated with the lowest Cd concentrations of 100 and 300 μM (Fig. 5l). The only exceptional genes related with ABA biosynthesis exhibiting a differential expression pattern in Cd-induced stress in comparison to Ni treatment were CpNCED2, which expression was not affected under Cd stress situation (Fig. 5h) and CpNCED4, that exhibited a repression under all Cd concentrations between 81.2 and 87.4% in comparison to control (Fig. 5j). Thus, most of the analyzed genes involved in ABA production and catabolism were upregulated under stress induced by Cd, with the only exception of those belonging to NCED family.
3.6 Data Integration and Visualization
The data obtained from total biomass analysis, joint to the expression of JA- and ABA-related genes was integrated in a PCA analysis and a heatmap to improve result visualization (Fig. 6). PCA analysis explained a 64.4 of the total variation, distributing the different groups according to the applied TME ion in the PCA plot (Fig. 6b), appearing the control group in the right upper part of the graph (blue dot), the groups from Cd-stressed plants in the middle-lower part (red dots) and those from Ni-treated plants in the left upper side (green dots). Integrating this information with the provided from the loading plot (Fig. 6a), the importance of the overexpression CpAAO3 and CpAOS2 becomes evident to differentiate the Cd-stressed groups, whereas CpJAR1, CpNCED4, and CpZEP were the most relevant genes responsible for the differences observed in the groups treated with Ni.
Comparative analyses of the different parameters analyzed in this work in C. pepo plants subjected to TME stress induced by Ni or Cd at different concentrations. a Loading plot of principal component analysis, with blue arrows referring to physiological and phenotypical data, red arrows to genes related with JA biosynthesis, and green arrows related with genes involved in ABA biosynthesis; b principal component analysis, where blue circles refer to control, green circles refer to Ni-stressed plants, and red circles refer to Cd-stressed plants; and c heatmap of the gene expression analysis for the different groups, where green colors refer to repressed genes and red colors to overexpressed genes
Additionally, the performed heatmap (Fig. 6c) also allows to easily point the common genetic response observed in plants treated with Ni or Cd, as it has been described previously in the “Results.”
4 Discussion
Phytohormones such as ABA and JA play essential roles in the regulation of the plant responses to stress. The importance of these two phytohormones has been widely studied in plants subjected to TME-induced stress. Generally, their concentration is increased under this hazardous condition, and it is related to a better tolerance to high concentration of toxic metal ions (Bücker-Neto et al., 2017; Kim et al., 2021). Additionally, these two plant hormones, ABA and JA have a synergistic interaction under TME-induced stress (Kumar et al., 2022), and their exogenous application may improve plant tolerance to metal stress (Ali et al., 2018; Zhao et al., 2023). Consequently, the study of the genetic regulation of their synthesis is important for the selection of candidate genes to be used in breeding programs, which may conduct to the obtention of new varieties with an enhanced capacity to naturally synthesize these plant signaling compounds and contributing to an enhanced crop tolerance to the harmful situation caused by TME exposition.
The leaf damage observed in C. pepo plants exposed to Ni revealed the higher sensitivity of this plant species to Ni in comparison to Cd, which was correlated with the decrease of leaf and root biomass at lower concentrations of Ni in comparison to Cd (Figs. 1 and 2). This higher toxicity of Ni has been already described in earlier studies with other crops as tomato, which also exhibits a decrease in the root and leaf length and biomass production (Badawy et al., 2022), or maize, which biomass diminished more in the presence of Ni than in Cd-treated plants (Pavlovkin et al., 2016). In this line, the capacity of C. pepo plants to avoid the translocation of Cd to the aerial organs (differently to that observed with Ni treatments), would support the idea of a higher tolerance to Cd of this plant species. This is supported by the different accumulation pattern of these metal ions, because while Ni was distributed at similar concentrations in leaves and roots, Cd was mainly accumulated in roots, resulting in a lower translocation factor to the aerial tissues. This result contradicts the observations of other authors in tomato, where both elements, Cd and Ni, were accumulated at similar levels in both tissues (Badawy et al., 2022).
Up to date, few works regarding the genetic expression of C. pepo in response to stress conditions have been reported, and they are focused on the study of genes related to defense proteins or phytohormones such as salicylic acid and ethylene in response to Phytophthora capsici infection (Ayala-Doñas et al., 2021) or the study of the WRKY transcription factor family in C. pepo plants subjected to drought or salt stress conditions (Bankaji, Sleimi, et al., 2019). A lack of information exists in the case of TME-induced stress. In this work, we have identified 12 genes involved in JA and ABA biosynthesis in this species, and their relative expression has been analyzed in response to different concentrations of Ni or Cd. The obtained results support that gene expression patterns are almost parallel, independently of the toxic metal applied, revealing a similar genetic response to both elements.
It is widely accepted that ABA helps plants to prevent and mitigate the harmful effect of TME toxicity, and external applications of this phytohormone have been used for the prevention of TME damage in different crops (reviewed in Kumar et al., 2022). Previous works with zucchini plants treated with Cd did not detect differences in the ABA leaf content but reported an increase in the endogenous concentrations of this phytohormone in roots and fruits (Labidi et al., 2021). This information could imply that in C. pepo plants subjected to Cd, ABA is synthesized in leaves and rapidly transported to roots and fruits, justifying the overexpression of genes such as CpZEP or CpAAO3, crucial for the regulation of ABA synthesis. This hypothesis agrees with the higher shoot carotenoid content, and recent advances that suggest that aerial organs are determinant in the stress perception and signaling to the root in the regulation of ABA biosynthesis (Manzi et al., 2015) Previous works with rice plants subjected to Cd-induced stress have concluded the importance of AAO3 upregulation in the tolerance to this metal ion, being highly induced in CBL-interacting protein kinase 11 (CIPK11) overexpressing lines (Gu et al., 2021), which may suggest that in our work, C. pepo plants would tolerate better TME-induced stress at lower concentrations (100 and 300 μM), but not at higher doses of 500 μM, where the expression of CpAAO3 is repressed. The enhanced expression of CpCYP707A2 could lead to ABA hydroxylation to phaseic and dihydrophaseic acid, two catabolites that are often accumulated in plants under abiotic stress situations (Hewage et al., 2020). Additionally, an increase in ABA biosynthesis-related genes (as occurs in this work with CpZEP in plants treated with TME) is usually correlated with an enhanced overexpression of genes related with the catabolism of this phytohormone to regulate precisely and rapidly its content (Long et al., 2019). Regarding NCED family, previous studies have concluded that there is an upregulation of StNCED1 in roots of potato treated with Cd (Stroiński et al., 2013), or different members of this family in citrus plants treated with Cd (Gavassi et al., 2021). However, the results obtained from this work point out that there is a downregulation of CpNCED1, CpNCED2, and CpNCED3 in most of the studied situations, whereas CpNCED4 is upregulated in plants stressed with Ni but not in those treated with Cd. In this case, more investigation would be needed, since the efficiency regulating ABA biosynthesis of the distinct members of NCED family could be different among species and may be lower in C. pepo, where the regulation of the expression of some NCED members could be less critical for ABA biosynthesis.
Regarding gene expression of JA biosynthesis and catabolism pathway, the importance of the coexistence of various bioactive forms as OPDA, JA, or the conjugates of JA with different amino acids as JA-Ile, JA-Leu, or JA-Val must be considered (Ruan et al., 2019). Previous works have reported the importance of JA pathway under Cd stress, as occurs in tomato, where spr2 JA-deficient mutant is more sensitive to this toxic ion (Zhao et al., 2016), although other studies with adult C. pepo plants subjected to TME-induced stress have reported that JA content is not affected by Cd treatment in comparison to control (Labidi et al., 2021). In fact, it has been demonstrated that JA is capable to regulate photosynthesis and phytochelatin content in Brassica juncea plants subjected to Pb (Agnihotri & Seth, 2020), justifying that the protective role of JA could be more evident in adult plants since they are more photosynthetically active than seedlings.
In response to other toxic cations different to Cd or Ni, the literature studying the response of phytohormone biosynthesis-related genes is scarce, maybe due to the lower importance of other TME elements in comparison to Cd or Ni. Previous works regarding the overexpression of some of these genes have conducted to the obtention of TME tolerant plants, as Arabidopsis plants overexpressing cotton AOC1 gene (Wang et al., 2015) or tobacco plants overexpressing wheat AOS gene (Liu et al., 2014) have contributed to the obtainment of lines tolerant to Cu or Zn, respectively. Additionally, works with rice plants subjected to As (V)-induced stress have reported an upregulated expression of the ABA biosynthesis genes (Huang et al., 2012).
In our work, the genes upstream in the JA biosynthetic pathway CpLOX2 and CpAOS1, involved in the conversion from linolenic acid into OPDA were generally repressed, but CpAOS2 was upregulated under Cd-induced stress. Although the AOS family is typically upregulated during abiotic stress conditions, the accumulation of JA in the signaling of abiotic stress is transient, and it is likely that the downregulation of genes related to JA biosynthesis occurs after reaching the maximum concentration of jasmonates. (De Ollas et al., 2013). Meanwhile, the gene CpOPR3 responsible for the OPDA conversion into JA exhibited a different pattern depending on the applied metal and doses, and previous works have reported that it is highly involved in the activation of heavy metal stress adaptive pathways in other crops as rice (Di et al., 2021). Finally, CpJAR1, responsible of the conjugation of JA to JA-Ile, was upregulated under both stressful situations, which could be impaired with a conversion of JA to the conjugated form JA-Ile, but further investigations would be needed. Integrating the expression from JA-related genes, it seems that this pathway would be focused on the regulation of JA-Ile production, considered the most bioactive jasmonate and could be crucial for the tolerance to this stressful situation, since strong reductions of this molecule have been reported in maize subjected to TME-induced stress (Matayoshi et al., 2022).
5 Conclusions
Both phytohormones, ABA and JA, play an essential role in the signaling of TME-induced stress in C. pepo plants, being the response of the main genes involved in their biosynthesis similar between Ni and Cd treatments. In addition, ABA-related genes CpAAO3, CpZEP, and CpNCED4, as well as JA biosynthesis related genes CpLOX2, CpOPR3, CpAOS2, and CpJAR1, could play an essential role in the phytohormone response under TME stress conditions. These genes could be used as markers for future breeding programs of C. pepo plants focused on the obtention of new varieties with enhanced basal levels of these two phytohormones, contributing to an enhanced tolerance to TME-induced stress.
Data Availability
Data will be made available on reasonable request.
Change history
08 December 2023
The Orcids, Crossmark and Published Online Date has been updated.
References
Agnihotri, A., & Seth, C. S. (2020). Does jasmonic acid regulate photosynthesis, clastogenecity, and phytochelatins in Brassica juncea L. in response to Pb-subcellular distribution? Chemosphere, 243, 125361. https://doi.org/10.1016/j.chemosphere.2019.125361
Ali, E., Hussain, N., Shamsi, I. H., Jabeen, Z., Siddiqui, M. H., & Jiang, L. X. (2018). Role of jasmonic acid in improving tolerance of rapessed (Brassica napus L.) to Cd toxicity. Journal of Zheijang University-SCIENCE B, 19, 130–146. https://doi.org/10.1631/jzus.B1700191
Ayala-Doñas, A., Gómez, P., & De Cara-García, M. (2021). Gene expression in Cucurbita spp. root and crown during Phytophthora capsici infection. Plants, 10, 2718. https://doi.org/10.3390/plants10122718
Badawy, I. H., Hmed, A. A., Sofy, M. R., & Al-Mokadem, A. Z. (2022). Alleviation of cadmium and nickel toxicity and phyto-stimulation of tomato plant L. by endophytic Micrococcus luteus and Enterobacter cloacae. Plants, 11, 2018. https://doi.org/10.3390/plants11152018
Bankaji, I., Pérez-Clemente, R. M., Caçador, I., & Sleimi, N. (2019). Accumulation potential of Atriplex halimus to zinc and lead combined with NaCl: Effects on physiological parameters and antioxidant enzymes activities. South African Journal of Botany, 123, 51–61. https://doi.org/10.1016/j.sajb.2019.02.011
Bankaji, I., Sleimi, N., Vives-Peris, V., Gómez-Cadenas, A., & Pérez-Clemente, R. M. (2019). Identification and expression of the Cucurbita WRKY transcription factors in response to water deficit and salt stress. Scientia Horticulturae (Amsterdam), 256, 108562. https://doi.org/10.1016/j.scienta.2019.108562
Begum, W., Rai, S., Banerjee, S., Bhattacharjee, S., Mondal, M. H., Bhattarai, A., & Saha, B. (2022). A comprehensive reciew on the sources, essentiality and toxicological profile of nickel. RSC Advances, 12, 9139–9153. https://doi.org/10.1039/d2ra00378c
Bouslimi, H., Ferreira, R., Dridi, N., Brito, P., Martins-Dias, S., Caçador, I., & Sleimi, N. (2021). Effects of barium stress in Brassica juncea and Cakile maritima: The indicator role of some antioxidant enzymes and secondary metabolites. Phyton – International Journal of Experimental Botany, 90, 145–158. https://doi.org/10.32604/phyton.2020.011752
Bücker-Neto, L., Paiva, A. L. S., Machado, R. D., Arenhart, R. A., & Margis-Pinheiro, M. (2017). Interactions between plant hormones and heavy metals responses. Genetics and Molecular Biology, 40, 373–386. https://doi.org/10.1590/1678-4685-gmb-2016-0087
Cesur, A., Cetin, I. Z., Cetin, M., Sevik, H., & Ozel, H. B. (2022). The use of Cupressus arizonica as a biomonitor of Li, Fe, and Cr pollution in Kastamonu. Water, Air, & Soil Pollution, 233, 193. https://doi.org/10.1007/s11270-022-05667-w
Cetin, M., Aljama, A. M. O., Alrabiti, O. B. M., Adiguzel, F., Sevik, H., & Cetin, I. Z. (2022). Using topsoil analysis to determine and map changes in Ni Co pollution. Water, Air, & Soil Pollution, 233, 293. https://doi.org/10.1007/s11270-022-05762-y
Chrysargyris, A., Höfte, M., Tzortzakis, N., Petropoulos, S. A., & Di Gioia, F. (2022). Editorial: Micronutrients: The borderline between their beneficial role and toxicity in plants. Frontiers in Plant Science, 13, 840624. https://doi.org/10.3389/fpls.2022.840624
de Ollas, C., Hernando, B., Arbona, V., & Gómez-Cadenas, A. (2013). Jasmonic acid transient accumulation is needed for abscisic acid increase in citrus roots under drought stress conditions. Physiologia Plantarum, 147, 296–306. https://doi.org/10.1111/j.1399-3054.2012.01659.x
Di, X., Zheng, F., Norton, G. J., Beesley, L., Zhang, Z., Lin, H., Zhi, S., Liu, X., & Ding, Y. (2021). Physiological responses and transcriptome analyses of upland rice following exposure to arsenite and arsenate. Environmental and Experimental Botany, 183, 104366. https://doi.org/10.1016/j.envexpbot/2020.104366
Fabiano, C. C., Tezotto, T., Favarin, J. L., Polacco, J. C., & Mazzafera, P. (2015). Essentiality of nickel in plants: A role in plant stresses. Frontiers in Plant Science, 6, 754. https://doi.org/10.3389/fpls.2015.00754
Galal, T. M. (2016). Health hazards and heavy metals accumulation by summer squash (Cucurbita pepo L.) cultivated in contaminated soils. Environmental Monitoring and Assessment, 188, 434. https://doi.org/10.1007/s10661-016-5448-3
Gavassi, M. A., Silva, G. S., da Silva, C. M., Thompson, A. J., Macleod, K., Oliveira, P. M. R., Cavalheiro, M. F., Domingues, D. S., & Haberman, G. (2021). NCED expression is related to increased ABA biosynthesis and stomatal closure under aluminum stress. Environmental and Experimental Botany, 185, 104404. https://doi.org/10.1016/j.envexpbot.2021.104404
Gu, S., Wang, X., Bai, J., Wei, T., Sun, M., Zhu, L., Wang, M., Zhao, Y., & Wei, W. (2021). The kinase CIPK11 functions as a positive regulator in cadmium stress response in Arabidopsis. Gene, 772, 145372. https://doi.org/10.1016/j.gene.2020.145372
Hewage, K. A. H., Yang, J. F., Wang, D., Hao, G. F., Yang, G. F., & Zhu, J. K. (2020). Chemical manipulation of abscisic acid signaling: A new approach to abiotic and biotic stress management in agriculture. Advanced Science, 7, 2001265. https://doi.org/10.1002/advs.202001265
Hoagland, D., & Arnon, D. (1938). The water-culture method for growing plants without soil. Californian Agricultural Experimental Station, C347, 1–39.
Huang, T. L., Nguyen, Q. T. T., Fu, S. F., Lin, C. Y., Chen, Y. C., & Huang, H. J. (2012). Transcriptomic changes and signalling pathways induced by arsenic stress in rice roots. Plant Molecular Biology, 80, 587–608. https://doi.org/10.1007/s11103-012-9969-z
Kar, M. (2020). Expression patterns of oxidative stress-related genes of Cucurbita pepo and relation to cellular H2O2 under short-term heavy metal stress. Journal of Integrated Science and Technology, 10, 2952–2961. https://doi.org/10.21597/jist.729977
Kim, H., Seomun, S., Yoon, Y., & Jang, G. (2021). Jasmonic acid in plant abiotic stress tolerance and interaction with abscisic acid. Agronomy, 11, 1886. https://doi.org/10.3390/agronomy11091886
Koressaar, T., & Remm, M. (2007). Enhancements and modifications of primer design program Primer3. Bioinformatics, 23, 1289–1291. https://doi.org/10.1093/bioinformatics/btm091
Kouki, R., Ayachi, R., Ferreira, R., & Sleimi, N. (2021). Behavior of Cucumis sativus L. in presence of aluminum stress: Germination, plant growth and antioxidant enzymes. Food Science & Nutrition, 9, 3280–3288. https://doi.org/10.1002/fsn3.2294
Kouki, R., Dridi, N., Vives-Peris, V., Gómez-Cadenas, A., Caçador, I., Pérez-Clemente, R. M., & Sleimi, N. (2023). Appraisal of Abelmoschus esculentus L. response to aluminum and barium stress. Plants, 12, 179. https://doi.org/10.3390/plants12010179
Kubier, A., Wilkin, R. T., & Pichler, T. (2019). Cadmium in soils and groundwater. Applied Geochemistry, 108, 1–16. https://doi.org/10.10167/j.apgeochem.2019.104388
Kumar, S., Shah, S. H., Vimala, Y., Jatav, H. S., Ahmad, P., Chen, Y., & Siddique, K. H. M. (2022). Abscisic acid: Metabolism, transport, crosstalk with other plant growth regulators, and its role in heavy metal stress mitigation. Frontiers in Plant Science, 13, 972856. https://doi.org/10.3389/fpls.2022.972856
Labidi, O., Vives-Peris, V., Gómez-Cadenas, A., Pérez-Clemente, R. M., & Sleimi, N. (2021). Assessing of growth, antioxidant enzymes, and phytohormone regulation in Cucurbita pepo under cadmium stress. Food Science and Nutrition, 9, 2021–2031. https://doi.org/10.1002/fsn3.2169
Liu, H. H., Wang, Y. J., Wang, S. P., Li, H. J., & Xin, Q. G. (2014). Improved zinc tolerance of tobacco by transgenic expression of an allene oxide synthase gene from hexaploid wheat. Acta Physiologiae Plantarum, 36, 2433–2440. https://doi.org/10.1007/s11738-014-1616-7
Long, H., Zheng, Z., Zhang, Y., Xing, P., Wan, X., Zheng, Y., & Li, L. (2019). An abscisic acid (ABA) homeostasis regulated by its production, catabolism and transport in peanut leaves in response to drought stress. PLoS ONE, 14, e0213963. https://doi.org/10.1371/journal.pone.0213963
Manzi, M., Lado, J., Rodrigo, M. J., Zacarías, L., Arbona, V., & Gómez-Cadenas, A. (2015). Root ABA accumulation in long-term water-stressed plants is sustained by hormone transport from aerial organs. Plant and Cell Physiology, 56, 2457–2466. https://doi.org/10.1093/pcp/pcv161
Matayoshi, C. L., Pena, L. B., Arbona, V., Gómez-Cadenas, A., & Gallego, S. M. (2022). Biochemical and hormonal changes associated with root growth restriction under cadmium stress during maize (Zea mays L.) pre-emergence. Plant Growth Regulation, 96, 269–281. https://doi.org/10.1007/s10725-021-00774-w
Montero-Pau, J., Blanca, J., Bombarely, A., Ziarsolo, P., Esteras, C., Martí-Gómez, C., Ferriol, M., Gómez, P., Jamilena, M., Mueller, L., Picó, B., & Cañizares, J. (2018). De novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnology Journal, 16, 1161–1171. https://doi.org/10.1111/pbi.12860
Okereafor, U., Makhatha, M., Mekuto, L., Uche-Okereafor, N., Sebola, T., & Mavumengwana, V. (2020). Toxic metal implications on agricultural soils, plants, animals, aquatic life and human health. International Journal of Environmental Research and Public Health, 17, 1–24. https://doi.org/10.3390/ijerph17072204
Pavlovkin, J., Artiushenko, T., Syshchykov, D., Fiala, R., Repka, V., & Čiamporová, M. (2016). Physiological responses to cadmium, nickel and their interaction in the seedlings of two maize (Zea mays L.) cultivars. Agriculture (Pol’nohospodárstvo), 62, 127–137. https://doi.org/10.1515/agri-2016-0013
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research, 29, 2002–2007. https://doi.org/10.1093/nar/29.9.e45
Pfaffl, M. W., Horgan, G. W., & Dempfle, L. (2002). Relative expression software tool ( REST © ) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research, 30, 1–10. https://doi.org/10.1093/nar/30.9.e36
Rahi, A. A., Younis, U., Ahmed, N., Ali, M. A., Fahad, S., Sultan, H., Zarei, T., Danish, S., Taban, S., El Enshasy, H. A., Tamunaidu, R., Alotaibi, J. M., Alharbi, S. A., & Datta, R. (2022). Toxicity of cadmium and nickel in the context of applied activated carbon biochar for improvement in soil fertility. Saudi Journal of Biological Sciences, 29, 743–750. https://doi.org/10.1016/j.sjbs.2021.09.035
Ruan, J., Zhou, Y., Zhou, M., Yan, J., Khurshid, M., Weng, W., Cheng, J., & Zhang, K. (2019). Jasmonic acid signaling pathway in plants. International Journal of Molecular Sciences, 20, 2479. https://doi.org/10.3390/ijms20102479
Saini, S., Kaur, N., & Pati, P. K. (2021). Phytohormones: Key players in the modulation of heavy metal stress tolerance in plants. Ecotoxicology and Environmental Safety, 223, 112578. https://doi.org/10.1016/j.ecoenv.2021.112578
Sghaier, D., Bankaji, I., Pedro, S., Caçador, I., & Sleimi, N. (2019). Photosynthetic behaviour and mineral nutrition of Tamarix gallica cultivated under aluminium and NaCl combined stress. Phyton-International Journal of Experimental Botany, 88, 239–252. https://doi.org/10.32604/phyton.2019.06887
Shukla, A., Srivastava, S., & Suprasanna, P. (2017). Genomics of metal stress-mediated signalling and plant adaptive responses in reference to phytohormones. Current Genomics, 18, 512–522. https://doi.org/10.2174/1389202918666170608093327
Sleimi, N., Kouki, R., Hadj Ammar, M., Ferreira, R., & Pérez-Clemente, R. (2021). Barium effect on germination, plant growth, and antioxidant enzymes in Cucumis sativus L. plants. Food Science & Nutrition, 9, 2086–2094. https://doi.org/10.1002/fsn3.2177
Stroiński, A., Giżewska, K., & Zielezińska, M. (2013). Abscisic acid is required in transduction of cadmium signal to potato roots. Biologia Plantarum, 57, 121–127. https://doi.org/10.1007/s10535-012-0135-x
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., & Rozen, S. G. (2012). Primer3-new capabilities and interfaces. Nucleic Acids Research, 40, e115. https://doi.org/10.1093/nar/gks596
Vives-Peris, V., Marmaneu, D., Gómez-Cadenas, A., & Pérez-Clemente, R. M. (2018). Characterization of Citrus WRKY transcription factors and their responses to phytohormones and abiotic stresses. Biologia Plantarum, 62, 33–44. https://doi.org/10.1007/s10535-017-0737-4
Wang, Y., Liu, H., & Xin, Q. (2015). Improvement of copper tolerance of Arabidopsis by transgenic expression of an allene oxide cyclase gene, GhAOC1, in upland cotton (Gossypium hirsutum L.). The Crop Journal, 3, 343–352. https://doi.org/10.1016/j.cj.2015.02.004
Wasternack, C., & Song, S. (2017). Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. Journal of Experimental Botany, 68, 1303–1321. https://doi.org/10.1093/jxb/erw443
Xun, E., Zhang, Y., Zhao, J., & Guo, J. (2017). Translocation of heavy metals from soils into floral organs and rewards of Cucurbita pepo: Implications for plant reproductive fitness. Ecotoxicological and Environmental Safety, 145, 235–243. https://doi.org/10.1016/j.ecoenv.2017.07.045
Zhang, W., Zhang, Z., Song, J., Yue, S., & Yang, H. (2019). Cd2+ uptake inhibited by MhNCED3 from Malus hupehensis alleviates Cd-induced cell death. Environmental and Experimental Botany, 166, 103802. https://doi.org/10.1016/j.envexpbot.2019.103802
Zhao, S., Ma, Q., Xu, X., Li, G., & Hao, L. (2016). Tomato jasmonic acid-deficient mutant spr2 seedling response to cadmium stress. Journal of Plant Growth Regulation, 35, 603–610. https://doi.org/10.1007/s00344-015-9563-0
Zhao, Y., Wang, J., Huang, W., Zhang, D., Wu, J., Li, B., Li, M., Liu, L., & Yan, M. (2023). Abscisic-acid-regulated responses to alleviate cadmium toxicity in plants. Plants, 12, 1023. https://doi.org/10.33907/plants12051023
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This investigation has been economically supported by MCIN/AEI/10.13039/501100011033 and the European Union (grant number PID2022-137825OB-I00), and by Universitat Jaume I (grant numbers UJI-A2021-10 and POSDOC/2020/18).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
Sup. Tab. 1 Nutrient solution used for watering the plants during the experiment. (DOCX 13 kb)
ESM 2
Sup. Tab. 2 List of accessions and designed primers of the selected C. pepo genes. (DOCX 15 kb)
ESM 3
Sup. Fig. 1 Jasmonic acid biosynthesis pathway in plants. (JPG 1509 kb)
ESM 4
Sup. Fig. 2 Abscisic acid biosynthesis pathway in plants. (JPG 758 kb)
ESM 5
Sup. Fig. 3 Calibration curves used for the quantification of Cd (A) and Ni (B). (JPG 22 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Labidi, O., Kouki, R., Pérez-Clemente, R.M. et al. Involvement of Abscisic Acid and Jasmonic Acid Biosynthesis-related Genes in Cucurbita pepo L. Tolerance to Trace Metal Stress. Water Air Soil Pollut 234, 745 (2023). https://doi.org/10.1007/s11270-023-06763-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-023-06763-1