Abstract
Designing a single domain generalization (DG) framework that generalizes from one source domain to arbitrary unseen domains is practical yet challenging in medical image segmentation, mainly due to the domain shift and limited source domain information. To tackle these issues, we reason that domain-adaptive classifier learning and domain-agnostic feature extraction are key components in single DG, and further propose an adaptive infinite prototypes (InfProto) scheme to facilitate the learning of the two components. InfProto harnesses high-order statistics and infinitely samples class-conditional instance-specific prototypes to form the classifier for discriminability enhancement. We then introduce probabilistic modeling and provide a theoretic upper bound to implicitly perform the infinite prototype sampling in the optimization of InfProto. Incorporating InfProto, we design a hierarchical domain-adaptive classifier to elasticize the model for varying domains. This classifier infinitely samples prototypes from the instance and mini-batch data distributions, forming the instance-level and mini-batch-level domain-adaptive classifiers, thereby generalizing to unseen domains. To extract domain-agnostic features, we assume each instance in the source domain is a micro source domain and then devise three complementary strategies, i.e., instance-level infinite prototype exchange, instance-batch infinite prototype interaction, and consistency regularization, to constrain outputs of the hierarchical domain-adaptive classifier. These three complementary strategies minimize distribution shifts among micro source domains, enabling the model to get rid of domain-specific characterizations and, in turn, concentrating on semantically discriminative features. Extensive comparison experiments demonstrate the superiority of our approach compared with state-of-the-art counterparts, and comprehensive ablation studies verify the effect of each proposed component. Notably, our method exhibits average improvements of 15.568% and 17.429% in dice on polyp and surgical instrument segmentation benchmarks.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Deep neural networks (DNNs) have shown great performance and promise for broad applications, such as disease diagnosis (Chen & Xia, 2021) and medical image segmentation (Zhou et al., 2019), under the condition that the training and test data share similar distribution. However, this condition may not always hold true in real-world scenarios, and data distribution shifts caused by data corruptions (Zhao et al., 2020b), domain mismatches (Choi et al., 2021) or adversarial attacks (Wang et al., 2021b) may lead to performance degradation of DNNs. This performance degeneration hampers the generalizability and deployment of DNNs in real-world applications, where training and test data are usually drawn from different data distributions (Choi et al., 2021; Liu et al., 2021b), such as various medical centers and medical imaging devices.
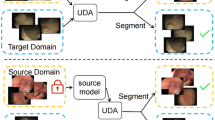
The data distribution discrepancy between training data (source domain) and test data (target domain), also known as domain shift (Fig. 1a), has been intensively studied in domain adaptation (DA) (Cheng et al., 2021; Li et al., 2021c; Liu et al., 2021a; b, Truong et al., 2023). Despite of the success in tackling domain discrepancy issue, DA tasks assume that the labeled or unlabeled target domain data is available, and this requirement limits the application of DA in practice since collecting data from every possible target domain and training DA models with each source-target pair are impractical (Choi et al., 2021). Hence, a more realistic yet challenging problem setting, i.e., domain generalization (DG) (Pandey et al., 2021), has attracted increasing attention. Different from DA tasks that require access to target domain data, DG aims to optimize DNNs with only single or multiple source domains and enhance the generalization capability to arbitrary unseen target domains. Single DG (Choi et al., 2021; Fan et al., 2021; Huang et al., 2021; Kundu et al., 2021; Li et al., 2021b; Qiao et al., 2020; Volpi et al., 2018; Zhang et al., 2020a; Zhao et al., 2020a) is more practical since the multi-source domain data required by multi-source DG (Ahmed et al., 2021; Bai et al., 2021; Dubey et al., 2021; He et al., 2021; Pandey et al., 2021; Su et al., 2023; Xu et al., 2021) task is expensive and labor-intensive to obtain, but the limited domain information raises challenges in single DG.
To tackle the single DG problem, existing methods utilizing data/domain augmentation (Huang et al., 2021; Kundu et al., 2021; Li et al., 2021b; Qiao et al., 2020; Volpi et al., 2018; Zhang et al., 2020a; Zhao et al., 2020a), adaptive normalization (Pan et al., 2019; Fan et al., 2021) and whitening transformation (Choi et al., 2021) techniques have shown promising results. Among these studies, adversarial domain augmentation (Li et al., 2021b; Qiao et al., 2020; Volpi et al., 2018; Zhang et al., 2020a; Zhao et al., 2020a) emerges as a strong baseline which synthesizes virtual domains in an adversarial way and retrains the deep model to learn the domain-invariant features with multiple virtual domain data. However, this heuristic adversarial training paradigm may not guarantee the safety and effectiveness of synthetic data (Li et al., 2021b), thus the optimized model is still vulnerable to severely domain-shifted target data (Qiao et al., 2020). An interesting yet seldom investigated problem then arises: can a model generalize from one source domain to many unseen target domains without data/domain augmentation? In other words, how to explicitly learn a domain-adaptive classifier and extract domain-agnostic features when there is only a single domain available for optimization? On the one hand, domain-adaptive classifier indicates the elasticity of mapping function for varying domains and aims to statistically adapt for each domain. On the other hand, domain-agnostic features are those representations that share across different domains and can preserve the generalization ability under domain shifts. In this work, instead of synthesizing new data/domains, we address the single DG problem from a brand-new perspective, where an adaptive infinite prototype scheme (InfProto) is built to explicitly enable the domain-adaptive classifier and domain-agnostic feature learning.
The domain-adaptive classifier has not yet been explored in previous single DG methods. Typically, they directly leverage the fully connected (FC) layer to perform classification (i.e., 1\(\times \)1 conv layer for image segmentation), where each category is represented by a single weight vector (Guo et al., 2021a). However, the optimized FC layer only involves the statistics estimated on source data, which inevitably results in performance drops on unseen target domain data. Moreover, the single weight vector in FC layer is insufficient to capture all characteristics of individual category (Guo et al., 2021b) and is limited in discriminability (Li et al., 2021a) because it merely considers the expectation information but neglects high-order statistics of feature representations (Chang et al., 2020; Guo et al., 2021b), such as variance information that embeds abundant semantic information (Han et al., 2019; Sohoni et al., 2020; Wang et al., 2021c; Yang et al., 2020). To cope with the aforementioned limitations of FC layer, we propose InfProto and incorporate it to design the domain-adaptive classifier. Specifically, InfProto infinitely samples class-conditional instance-specific prototypes to form the domain-adaptive classifier with flexible decision boundary, which can be adapted to each individual input data, as shown in Fig. 1b. Considering explicitly sampling infinite prototypes is infeasible, probabilistic modeling is introduced to implicitly perform the infinite prototype sampling, and an upper bound of the expected cross entropy (CE) loss is derived to inherently harness the impact of infinite prototypes. To ensure the robustness of the domain-adaptive classifier, mini-batch and dataset distributions are further incorporated to determine the range of sampled infinite prototypes (i.e., expectation and variance). Instances are grouped into mini-batches, which are then aggregated into datasets, establishing an inherent hierarchical relationship. With the infinite prototypes sampled from the hierarchical data distributions, the hierarchical domain-adaptive classifier is built, including instance-level and mini-batch-level domain-adaptive classifiers. It is worth noting that target domain data distributions are incorporated into the domain-adaptive classification in the inference phase, making the model generalizable and discriminable to arbitrary unseen domains.
To learn domain-agnostic features for single DG, existing methods (Huang et al., 2021; Li et al., 2021b; Qiao et al., 2020) optimize the deep model with multiple virtual domain data synthesized through the adversarial learning strategy. By eliminating domain shifts among synthesized data, the domain-specific information is suppressed, and domain-agnostic features are retained. However, they neglect the distribution shifts within a single source domain, which are induced by different instances exhibiting variant feature distributions (Fig. 1b). Based on the insight that domain-agnostic features can be theoretically distilled from a single source domain exhibiting distribution shifts (Creager et al., 2021), the instance-level infinite prototype exchange (ProtoEx) strategy is proposed. In particular, each instance within the source domain is regarded as a micro source domain, and distribution shifts among these micro source domains are minimized to derive domain-agnostic features. In order to explicitly minimize distribution shifts, instance-level infinite prototypes are exchanged among different instances within a mini-batch, and then the extracted feature map is encouraged to perform accurate segmentation via the classifier formed by exchanged prototypes. Through exchanging prototypes in ProtoEx strategy, the distribution information of two instances are swapped and constrained, increasing the robustness of model on variant micro source domains. According to the Law of Large Numbers, the statistical information of the desired data distribution should be more consistent with the mini-batch one in comparison to that of a single instance (Fig. 1c), and the corresponding mini-batch infinite prototypes represent the common knowledge sharing among variant micro source domains. Therefore, inheriting the spirit of ProtoEx, an instance-batch infinite prototype interaction (IBProto) strategy is further devised to regularize each instance, which should be precisely segmented with the classifier formed by mini-batch-level prototypes. Considering instance and mini-batch distributions should share similar representations if distribution shifts are eliminated, the consistency regularization (HieraCR) among outputs of the hierarchical domain-adaptive classifier is advanced to facilitate domain-agnostic feature learning. The contributions of this paper can be summarized in three aspects.
-
A brand-new paradigm is proposed for single DG, where the InfProto scheme is designed to form the domain-adaptive classifier and extract domain-agnostic features for promoting generalizability on unseen target domains.
-
A hierarchical domain-adaptive classifier with incorporated InfProto is designed to elasticize the segmentation model for varying domains and optimized by the derived upper bound with rigorous theoretical justification.
-
Domain-agnostic feature learning is guaranteed by the advanced ProtoEx, IBProto, HieraCR strategies, which explicitly minimize the distribution shifts among micro source domains within the single source domain.
-
Extensive experiments on polyp and instrument segmentation benchmarks demonstrate the proposed methods yield state-of-the-art results. Moreover, ablation studies are conducted to thoroughly verify the impact and effect of each proposed component.
2 Related Work
2.1 Image Segmentation
Image segmentation is the task of assigning a semantic label for each pixel within an image. It is an important task in computer vision and has a wide range of applications, such as medical image analysis (Guo et al., 2021b). Recently, the remarkable successes for image segmentation have mostly been achieved by deep convolutional neural networks, owing to their characterization ability for semantic representation (Minaee et al., 2021). Typically, most image segmentation networks follow the encoder-decoder design (Minaee et al., 2021). To overcome the low spatial resolution problem at the bottleneck, remedies such as skip connections (Ronneberger et al., 2015; Zhou et al., 2019), high-resolution representation maintenance (Fourure et al., 2017; Wang et al., 2020), and multi-scale fusion (Sagar & Soundrapandiyan, 2021; Zhao et al., 2017), are proposed. For example, HRNet (Wang et al., 2020) connects the high and low-resolution convolution branches in parallel and repeatedly exchanges the information between them, which results in the spatially precise representation feature extraction. Further improvements are achieved by harnessing context information, such as attention modules (Li et al., 2019, 2021d) or pyramid pooling (Zhao et al., 2017; Chen et al., 2018). Li et al. (2019) formulate the attention mechanism in an expectation-maximization manner, where a compact set of bases and attention maps are iteratively estimated. Unfortunately, the aforementioned deep models require large-scale densely annotated images, and the performance inevitably degrades when the well-optimized model is deployed on unseen domain data drawn from different data distributions (Li et al., 2021c; Pandey et al., 2021). Moreover, most image segmentation methods directly leverage the FC layer to perform classification, but the single weight vector in FC layer is insufficient to capture all characteristics of the individual category (Guo et al., 2021b), thereby limiting the discriminability (Li et al., 2021a).
In this paper, we mainly focus on medical image segmentation, which is more challenging than natural image segmentation due to the huge intra-class divergence and inter-class ambiguity. To get rid of annotating large-scale images, we explore the solution, termed InfProto, for single DG. Different from FC layer that represents each class with only a single weight vector, the proposed InfProto forms the domain-adaptive classifier with infinitely sampled instance-specific prototypes for promoting generalizability and facilitating the discriminability on unseen domains.
2.2 Domain Adaptation and Generalization
To deal with the domain shift problem and release the heavy annotation burden, domain adaptation (DA) has been extensively studied (Cheng et al., 2021; Kang et al., 2020; Kim & Byun, 2020; Li et al., 2021c; Liu et al., 2021a; Truong et al., 2023). Researchers explicitly characterize and narrow down the distribution discrepancy with the co-occurrence of source and target domains during training. Consequently, the optimized models are generalized well on the target domain even though the corresponding annotations are absent (Liu et al., 2021b). One popular line of DA approaches is learning domain-agnostic or domain-invariant representations by diminishing domain divergence (Liu et al., 2021a). The domain-agnostic feature follows the same distribution regardless of the domain discrepancy, and source domain features would resemble target domain characteristics (Zhang et al., 2019). Thus, the deep model optimized on source data can retain the generalizability for unlabeled target domain (Zhang et al., 2019). The domain-agnostic representation learning is achieved by minimizing a statistic metric (Kang et al., 2020) or adversarial learning (Cheng et al., 2021; Kim & Byun, 2020). Nevertheless, these methods require the co-occurrence of both domains during adaptation, which may be violated in real-world scenarios.
Unlike DA, which requires access to the target domain data during training, domain generalization (DG) exploits the knowledge from single or multiple seen source domains to learn a model that generalizes well on arbitrary unseen target domains. Herein, we explore the task of single DG (Ahmed et al., 2021; Bai et al., 2021; Dubey et al., 2021; He et al., 2021; Pandey et al., 2021; Su et al., 2023; Xu et al., 2021), which is more practical since the multi-source domain data required by multi-source DG (Choi et al., 2021) task is expensive and labor-intensive to obtain. In this case, the aforementioned domain-agnostic representation learning algorithms are infeasible to be directly applied to single DG tasks. Existing single DG methods usually utilize adaptive normalization (Fan et al., 2021; Pan et al., 2019), data/domain augmentation (Huang et al., 2021; Volpi et al., 2018; Zhao et al., 2020a; Qiao et al., 2020; Kundu et al., 2021; Li et al., 2021b; Zhang et al., 2020a) and whitening transformation (Choi et al., 2021) techniques. Switchable whitening (SW) (Pan et al., 2019) automatically switches among various normalization and whitening methods, hence, style information is removed for generalization enhancement. Li et al. (2021b) synthesize virtual domains in an adversarial way and leverage contrastive learning to learn the domain-agnostic features from multiple virtual domain data. Kundu et al. (2021) augment multiple novel domains to facilitate domain-agnostic representation learning. Choi et al. (2021) leverage feature covariance to disentangle the domain-invariant content from the domain-specific style and selectively remove the style representations causing domain shift.
In contrast to previous arts, we propose the hierarchical domain-adaptive classifier based on InfProto, which elasticizes for varying domains by incorporating statistical information of different domains. To our best knowledge, this is the first study to design domain-adaptive classifier for single DG. Moreover, considering that the heuristic adversarial training paradigm for domain augmentation (Huang et al., 2021; Li et al., 2021b; Qiao et al., 2020; Zhao et al., 2020a) may not guarantee the safety and effectiveness of synthetic data, we regard each instance within the single source domain as a micro source domain, and minimize distribution shifts among these micro source domains to derive domain-agnostic representations.
2.3 Prototype Learning
Since the huge variation between samples within the same category would result in a widely spread feature distribution, the FC layer is insufficient to represent all characteristics of individual categories (Han et al., 2019; Guo et al., 2021b). Recently, multi-prototype based methods have been developed to capture more characteristics of the individual category (Chang et al., 2020; Guo et al., 2021b; Han et al., 2019; Li et al., 2021a; Liu et al., 2020; Xu et al., 2020; Yang et al., 2020), thereby shaping a more accurate decision boundary. Han et al. (2019) propose self-learning with multi-prototypes to get a better representation and address the noisy label problem, where an extra partitioning scheme is devised to select the representative features as the multiple prototypes. Li et al. (2021a) present a flexible prototypical learning approach for few-shot segmentation, which incorporates superpixel to guide the extraction of multiple prototypes. However, this kind of method usually requires attribute prior (Xu et al., 2020), superpixel information (Li et al., 2021a) or a predefined number of multiple prototypes (Chang et al., 2020; Guo et al., 2021b; Han et al., 2019; Liu et al., 2020; Yang et al., 2020), and improper settings may be not able to deal with large variations in feature distributions across different images. To address those limitations, the proposed InfProto represents the first effort that samples infinite prototypes according to instance-specific and class-conditional statistical information, thereby comprehensively encoding all characteristics of individual classes and acquiring flexible decision boundary for each domain data.
3 Adaptive Infinite Prototypes (Infproto)
As analyzed in Sect. 1, domain-adaptive classifier learning and domain-agnostic feature extraction are key components for single DG. In conventional single DG models, the fully connected (FC) layer represents each category by a single weight vector, which only involves the 1st-moment information (i.e., expectation) of the source domain. The single weight vector in FC layer is insufficient to encode the diverse representations in unseen domains due to the limited domain-adaptive ability and the neglected abundant information in high-order statistics of the data distribution. The class-conditional covariance matrix captures the variance of instances within the same class, which represents the class-conditional distribution and contains rich semantic information (Wang et al., 2021c). To design a domain-adaptive classifier and fully explore distribution variations, we incorporate statistical information of data distribution, including 1st-moment (i.e., \( \varvec{\mu }\)) and 2nd-moment (i.e., \( \varvec{\Sigma }\)). A naive and intuitive method to implement domain-adaptive classifier is to explicitly encode the variant feature representations of a category y with multiple prototypes \({\varvec{p}}_{y} \in {\mathbb {R}}^{K \times D}\), which are sampled from the class-conditional data distribution \({\varvec{c}}_{y} \sim {\mathcal {N}} ( \varvec{\mu }_{y}, \varvec{\Sigma }_{y})\). Notice that K denotes the number of prototypes within the category y. Then, the segmentation model is optimized by minimizing CE loss on mini-batch data. For simplification, in this subsection, we derive \({\mathcal {L}}_{CE}\) for a pixel with feature \({\varvec{f}}\) and label y through: \({\mathcal {L}}_{CE}= \frac{1}{K}\sum _{k=1}^{K}-\log \frac{e^{{\varvec{f}} {\varvec{p}}^{k}_{y} }}{\sum _{j=1}^{C} e^{{\varvec{f}} {\varvec{p}}_{j}^{k}}}\), where C indicates the number of categories. Different from the FC layer, we discard the bias term to encourage the model to focus more on intrinsic data characteristics rather than adjusting to noise or domain-specific biases in the source data, thereby enhancing the generalizability of model. Obviously, when K is large enough to cover all possible prototypes, the naive implementation is not computationally applicable, as the cardinality of the prototype set is enlarged to \(C\times K\).
To circumvent the issue of computational efficiency, we consider the situation where K enlarges to infinity, and the effect of K is hopefully absorbed via probabilistic modeling. Mathematically, as K enlarges to infinity, we derive a computable upper bound for the expected CE loss, yielding a highly efficient implementation. To be specific, in the situation of \(K \rightarrow \infty \), we compute the expectation of CE loss with all possible prototypes, and \({\mathcal {L}}_{CE}^{\infty }\) is derived by:
where \(\lambda \) is a positive hyperparameter to control the range of sampling infinite prototypes. Since the expectation of the desired CE loss in Eq. (1) is intractable, we derive a rigorous closed-form of upper bound for \({\mathcal {L}}_{CE}^{\infty }\), as given by the following proposition.
Proposition 1
Suppose that \({\varvec{p}}_{y} \sim {\mathcal {N}} \left( \varvec{\mu }_{y}, {\lambda } \varvec{\Sigma }_{y}\right) \), then we have an upper bound of \({\mathcal {L}}_{CE}^{\infty }\), given by
Proof
With the concave property of logarithmic function, the upper bound of \({\mathcal {L}}_{CE}^{\infty }\) can be derived from Jensen’s inequality \({\mathbb {E}}_{x} \log (X) \le \log {\mathbb {E}}_{x}[X]\), and we have the upper bound as:
Since \({\varvec{p}}\) follows the distribution of \({\mathcal {N}} ( \varvec{\mu }, \lambda \varvec{\Sigma })\), we have \({\varvec{f}} {\varvec{p}} \sim {\mathcal {N}} \left( {\varvec{f}} \varvec{\mu }, ~\lambda {\varvec{f}} \varvec{\Sigma } {\varvec{f}}^{\top }\right) \). By leveraging the moment-generating function \({\mathbb {E}}\left[ e^{t X}\right] =e^{t^{\top }\left( \varvec{\mu }+\frac{1}{2} \varvec{\Sigma } t\right) }\), where \(\quad X \sim {\mathcal {N}} ( \varvec{\mu }, \varvec{\Sigma })\), we obtain an easy-to-calculate upper bound \({\mathcal {L}}_{Inf}\):
\(\square \)
Illustration of the proposed InfProto-powered framework, the main components of which are the adaptive infinite prototypes scheme (Sect. 3), domain-adaptive classifier (Sect. 4.1), and domain-agnostic feature learning (Sect. 4.2). Note that the right part ‘learning domain-agnostic feature’ takes the input \(x^{1}\) for example. Only the black solid lines have gradient backpropagation
Proposition 1 derives a surrogate loss for the proposed InfProto. Instead of directly optimizing the model with the desired CE loss \({\mathcal {L}}_{CE}^{\infty }\), we can minimize its upper bound \({\mathcal {L}}_{Inf}\) in a much more efficient way. Therefore, the proposed InfProto can be simply plugged into the segmentation model as a robust loss function, which inherently incorporates the abundant information in 2nd-moment statistics and harnesses the impact of infinite prototypes to form the classifier. The derived upper bound can be efficiently optimized with the stochastic gradient descent algorithm.
with the proposed adaptive infinite prototypes scheme (InfProto), the diverse feature representations within each category can be comprehensively encoded. Incorporating data distribution of test instance, the formed classifier thus can adapt well to unseen domains in inference stage.
4 Infproto-Powered Adaptive Classifier and Agnostic Feature Learning
The proposed InfProto can be used to facilitate domain-adaptive classifier learning and domain-agnostic feature extraction. Specifically, the InfProto-powered framework for single DG is illustrated in Fig. 2. Our goal is to train a segmentation model using single source domain data to correctly segment the images from an arbitrary target domain unseen in the inference process.
During the training phase, given source domain data \({\mathcal {D}}_{s} = \left\{ \left( {\varvec{X}}, {\varvec{Y}}\right) ^{i} \right\} _{i=1}^{N}\), mini-batch images \(\{ {\varvec{X}}^{i} \}_{i=1}^{n} \in {\mathcal {D}}_{s}\) are fed into a feature extractor \({\mathcal {F}}\) to obtain the corresponding feature maps \(\{ {\varvec{f}}^{i} \in {\mathbb {R}}^{H\times W\times D} \}_{i=1}^{n}\), where H, W, D denote the height, width, channel of feature maps. Hence, class-conditional data distribution in instance level (i.e., image level) is denoted as multivariate Gaussian distribution \({\varvec{c}}^{i}_{y} = {\mathcal {N}} ( \varvec{\mu }^{i}_{y}, \varvec{\Sigma }^{i}_{y})\), where \(\varvec{\mu }^{i}_{y} \in {\mathbb {R}}^{D\times 1}\) and \(\varvec{\Sigma }^{i}_{y} \in {\mathbb {R}}^{D\times D}\) are the mean and covariance matrix estimated from the instance-level feature map in class y according to the segmentation ground truth \(\{ {\varvec{Y}}^{i} \}_{i=1}^{n} \in {\mathcal {D}}_{s}\). Then, class-conditional data distribution \({\varvec{c}}^{b}_{y} = {\mathcal {N}} ( \varvec{\mu }^{b}_{y}, \varvec{\Sigma }^{b}_{y})\) in mini-batch-level is estimated from feature maps within a batch, and dataset-level \({\varvec{c}}^{d}_{y} = {\mathcal {N}} ( \varvec{\mu }^{d}_{y}, \varvec{\Sigma }^{d}_{y})\) are momentum updated with the statistics in history batches.
To form the hierarchical domain-adaptive classifier (Sect. 4.1), we leverage previously calculated class-conditional statistics \({\varvec{c}}^{i}_{y}\), \({\varvec{c}}^{b}_{y}\), and \({\varvec{c}}^{d}_{y}\). Specifically, these statistics determine the range of infinitely sampling prototypes \({\varvec{p}}^{I}\) and \({\varvec{p}}^{B}\), which are used to build the hierarchical domain-adaptive classifier, including instance-level and mini-batch-level classifiers. The hierarchical domain-adaptive classifier produces the instance-level segmentation prediction \(P^{I}\) and the mini-batch-level result \(P^{B}\) for each input image \({\varvec{X}}\). To learn the domain-agnostic feature (Sect. 4.2), we regard each instance within the source domain as a micro source domain and intend to distill from distribution shifts in a single source domain. In particular, we exchange instance-level prototypes among instances within a mini-batch and obtain the infinite prototypes \({\varvec{p}}^{E}\), thereby deriving the corresponding segmentation prediction \(P^{E}\). All predictions \(P^{I}\), \(P^{E}\), \(P^{B}\) are constrained with the ground truth y through the derived upper bound of the expected cross entropy (CE) loss \({\mathcal {L}}_{Inf}\). Moreover, the consistency regularization \({\mathcal {L}}^{HieraCR}\) among outputs \(P^{I}\), \(P^{E}\), \(P^{B}\) is constrained to facilitate domain-agnostic feature learning.
During the inference phase, target domain images are passed through the well-trained \({\mathcal {F}}\) and source domain classifier to obtain pixel-wise confidence score. Then, confident pixels are utilized to derive target domain moments, which are further used to form the hierarchical domain-adaptive classifier for the target domain, so as to produce the final segmentation results [see Eq. (6]).
4.1 Learning Domain-Adaptive Classifier
To elasticize the segmentation model for varying domains, we incorporate InfProto into the hierarchical domain-adaptive classifier instead of merely using a standard FC layer that is fixed for all target images once optimized. The key insight is that adapting the classifier per domain can be achieved by adapting the classifier per instance because each domain can be considered a distribution of all instances in statistics. Therefore, the parameters within the proposed hierarchical domain-adaptive classifier are explicitly conditioned on different instances to facilitate the generalizability on unseen domains. In the implementation, we establish a multivariate Gaussian distribution for each class at the instance level. The instance-level data distribution for class y is \({\varvec{c}}^{i}_{y} = {\mathcal {N}} \left( \varvec{\mu }^{i}_{y}, \varvec{\Sigma }^{i}_{y}\right) \), which is leveraged to determinate the range of sampled infinite prototypes and further used to form the instance-level domain-adaptive classifier.
Considering the instance-level distribution may deviate severely, we then introduce the mini-batch-level data distributions to derive the mini-batch-level domain-adaptive classifier. Formally, the class-conditional data distribution in mini-batch level is \({\varvec{c}}^{b}_{y} = {\mathcal {N}} \left( \varvec{\mu }^{b}_{y}, \varvec{\Sigma }^{b}_{y}\right) \). The above-formed instance-level and mini-batch-level domain-adaptive classifiers are unified to build the hierarchical domain-adaptive classifier.
To enhance the robustness of the hierarchical domain-adaptive classifier, the dataset-level distribution is further incorporated and computed by consecutively aggregating statistics from all history mini-batches in an online fashion:
where (t) indicates the current iteration, and \((t-1)\) is the last iteration. \(n_{y}^{(t-1)}\) denotes the total number of training pixels within \(y^{th}\) class in all mini-batches, and \(m_{y}^{(t)}\) denotes the number of training pixels belonging to \(y^{th}\) class only included in current \(t^{th}\) mini-batch. Therefore, the hierarchical domain-adaptive classifier is composed of two sets of infinite prototypes sampled from \({\mathcal {N}} (\varvec{\mu }^{I}_{}, ~ \varvec{\Sigma }^{I}_{} )={\mathcal {N}} \left( \lambda _{\varvec{\mu }} \varvec{\mu }^{i}_{} + (1-\lambda _{\varvec{\mu }}) \varvec{\mu }^{d}_{}, ~ \lambda _{\varvec{\Sigma }} \varvec{\Sigma }^{i}_{} + (1-\lambda _{\varvec{\Sigma }}) \varvec{\Sigma }^{d}_{} \right) \), \(~{\mathcal {N}} (\varvec{\mu }^{B}_{}, \varvec{\Sigma }^{B}_{})={\mathcal {N}} (\lambda _{\varvec{\mu }} \varvec{\mu }^{b}_{} + (1-\lambda _{\varvec{\mu }}) \varvec{\mu }^{d}_{}, ~ \lambda _{\varvec{\Sigma }} \varvec{\Sigma }^{b}_{} +\) \((1-\lambda _{\varvec{\Sigma }}) \varvec{\Sigma }^{d}_{})\), where \(\lambda _{\varvec{\mu }}\), \(\lambda _{\varvec{\Sigma }}\) control contributions of instance-level, mini-batch-level and dataset-level data distributions. It is worth noting when some classes are missing in an instance image, in the instance-level domain-adaptive classifier, the corresponding infinite prototypes are sampled only from dataset-level distribution, i.e., \({\mathcal {N}} (\varvec{\mu }^{I}_{}, ~ \varvec{\Sigma }^{I}_{} )={\mathcal {N}} ( \varvec{\mu }^{d}_{}, ~ \varvec{\Sigma }^{d}_{} )\). This classifier is optimized by minimizing the upper bound of desired CE loss \({\mathcal {L}}_{Inf}\) [Eq. (4)] in the training stage.
During the inference phase, test target domain images are first fed into the well-trained \({\mathcal {F}}\) to obtain the feature \({\varvec{f}}^{t}\). Then, confident pixels whose maximum predicted probability is greater than 0.7 are utilized to calculate the target domain moments, i.e., \(\varvec{\mu }^{t}\) and \(\varvec{\Sigma }^{t}\), in both instance \({\varvec{c}}^{i}={\mathcal {N}} (\varvec{\mu }^{i}, ~ \varvec{\Sigma }^{i})\) and mini-batch levels \({\varvec{c}}^{b}={\mathcal {N}} (\varvec{\mu }^{b}, ~ \varvec{\Sigma }^{b})\), and the dataset-level distribution \({\varvec{c}}^{d}={\mathcal {N}} (\varvec{\mu }^{d}, ~ \varvec{\Sigma }^{d})\) is borrowed from the source domain to retain the semantic information. These statistics are leveraged to form the hierarchical domain-adaptive classifier for the target domain so as to obtain the final segmentation results via
When \({\varvec{p}}^{t}_{j}\) is sampled from target domain instance-level distribution \({\mathcal {N}} \left( \lambda _{\varvec{\mu }} \varvec{\mu }^{i}_{j} + (1-\lambda _{\varvec{\mu }}) \varvec{\mu }^{d}_{j}, ~ \lambda _{\varvec{\Sigma }} \varvec{\Sigma }^{i}_{j} + (1-\lambda _{\varvec{\Sigma }}) \varvec{\Sigma }^{d}_{j} \right) \), we can obtain the segmentation prediction \(P^{I}_{j}\). If \({\varvec{p}}^{t}_{j}\) is sampled from the target domain mini-batch data distribution \({\mathcal {N}} \left( \lambda _{\varvec{\mu }} \varvec{\mu }^{b}_{j} + (1-\lambda _{\varvec{\mu }}) \varvec{\mu }^{d}_{j}, ~ \lambda _{\varvec{\Sigma }} \varvec{\Sigma }^{b}_{j} + (1-\lambda _{\varvec{\Sigma }}) \varvec{\Sigma }^{d}_{j} \right) \), the segmentation result \(P^{B}_{j}\) is derived. The final prediction is computed through \(\arg \max _{j} (P^{I}_{j} + P^{B}_{j})\).
The proposed hierarchical domain-adaptive classifier simplifies the alignment between the source domain and unavailable target domains by elasticizing the model for varying domains, which largely eases the learning requirement. Moreover, the hierarchical domain-adaptive classifier is adjusted according to the domain-specific infinite prototypes, facilitating the generalizability on unseen domains in the inference stage.
4.2 Learning Domain-Agnostic Feature
To learn domain-agnostic feature, we make full use of distribution shifts within a single source domain to learn domain-agnostic features. In particular, we regard each instance within the source domain as a micro source domain, and minimize distribution shifts among these micro source domains to derive domain-agnostic features. Technically, we propose three complementary strategies, including instance-level infinite prototype exchange (ProtoEx, Sect. 4.2.1), instance-batch infinite prototype interaction (IBProto, Sect. 4.2.2) and consistency regularization (HieraCR, Sect. 4.2.3), to facilitate the domain-agnostic feature learning.
4.2.1 Infinite Prototype Exchange (ProtoEx)
The main idea of ProtoEX is to explicitly minimize the distribution shifts existing among micro source domains, so as to retain the domain-agnostic features. To this end, we randomly exchange the instance-level infinite prototypes among different instances within a batch and then encourage the extracted feature map to perform accurate segmentation via the classifier formed by the exchanged prototypes. This is achieved by minimizing \({\mathcal {L}}_{Inf}^{E}\), which is calculated via
where \({\mathcal {N}} \left( \varvec{\mu }^{E}_{j}, ~ \varvec{\Sigma }^{E}_{j} \right) \) is the class-conditional data distribution of another instance within the mini-batch. This exchange encourages micro source domains to have comparable feature distributions despite potential distribution shifts in source domain. In the meanwhile, the instance-level prediction is regularized by minimizing \({\mathcal {L}}_{Inf}^{I}\), which is derived by replacing \({\mathcal {N}} \left( \varvec{\mu }^{E}_{j}, ~ \varvec{\Sigma }^{E}_{j} \right) \) with \({\mathcal {N}} \left( \varvec{\mu }^{I}_{j}, ~ \varvec{\Sigma }^{I}_{j} \right) \) in Eq. (7). By simultaneously minimizing \({\mathcal {L}}_{Inf}^{I}\) and \({\mathcal {L}}_{Inf}^{E}\), the input image is constrained to obtain accurate prediction with varying classifiers formed from different statistics, thereby preserving domain-agnostic features and increasing the robustness of the segmentation model on variant micro source domains.
4.2.2 Instance-Batch Infinite Prototype Interaction (IBProto)
According to the Law of Large Numbers, the statistical information of the desired data distribution should be more consistent with the mini-batch one compared with that of a single instance. Moreover, the sampled mini-batch infinite prototypes represent the common knowledge shared among variant micro source domains. Therefore, we inherit the spirit of ProtoEx and further design the IBProto strategy, which enforces the classifier formed by mini-batch-level prototypes to segment each input instance precisely. In particular, the exchanged class-conditional data distribution \({\mathcal {N}} \left( \varvec{\mu }^{E}_{j}, ~ \varvec{\Sigma }^{E}_{j} \right) \) in Eq. (7) is replaced with the mini-batch-level one, i.e., \({\mathcal {N}} \left( \varvec{\mu }^{B}_{j}, ~ \varvec{\Sigma }^{B}_{j} \right) \), and thus we obtain the \({\mathcal {L}}_{Inf}^{B}\). Optimizing \({\mathcal {L}}_{Inf}^{B}\) ensures the data distribution of each micro source domain to pull towards the mini-batch distribution. Hence, the interaction between instance and batch further minimizes the distribution shifts within the source domain.
4.2.3 Consistency Regularization (HieraCR)
Considering the instance-level and mini-batch-level data distribution should share similar representations if distribution shifts are eliminated, we further devise the HieraCR strategy. According to Eq. (6), three predictions can be obtained, including \(P^{I}=\frac{e^{\varvec{{\varvec{f}}\mu }_{y}^{I} + \frac{1}{2}\lambda {\varvec{f}} \varvec{\Sigma }_{y}^{I} {\varvec{f}}^{\top }}}{\sum _{j=1}^{C} e^{\varvec{{\varvec{f}}\mu }_{j}^{I} + \frac{1}{2}\lambda {\varvec{f}} \varvec{\Sigma }_{j}^{I} {\varvec{f}}^{\top }}}\), \(P^{E}=\frac{e^{\varvec{{\varvec{f}}\mu }_{y}^{E} + \frac{1}{2}\lambda {\varvec{f}} \varvec{\Sigma }_{y}^{E} {\varvec{f}}^{\top }}}{\sum _{j=1}^{C} e^{\varvec{{\varvec{f}}\mu }_{j}^{E} + \frac{1}{2}\lambda {\varvec{f}} \varvec{\Sigma }_{j}^{E} {\varvec{f}}^{\top }}}\) and \(P^{B}=\frac{e^{\varvec{{\varvec{f}}\mu }_{y}^{B} + \frac{1}{2}\lambda {\varvec{f}} \varvec{\Sigma }_{y}^{B} {\varvec{f}}^{\top }}}{\sum _{j=1}^{C} e^{\varvec{{\varvec{f}}\mu }_{j}^{B} + \frac{1}{2}\lambda {\varvec{f}} \varvec{\Sigma }_{j}^{B} {\varvec{f}}^{\top }}}\). Due to the shifts among micro source domains and the discrepancy between source instance and mini-batch data, the resulting segmentation predictions are mutually inconsistent. To improve the stability and robustness of the segmentation net, we design HieraCR loss for optimization:
By regularizing the consistency among outputs of the hierarchical classifier, distribution shifts among micro source domains and mini-batch one are explicitly narrowed down, thus facilitating the domain-agnostic feature learning.
5 Experiment
5.1 Datasets
We evaluate the performance of the proposed InfProto frameworkFootnote 1 for single DG task on two challenging medical image segmentation tasks, including polyp segmentation and surgical instrument segmentation. Comprehensive experiments covering cross imaging device and cross surgical site scenarios are implemented to demonstrate the effectiveness of the proposed InfProto framework.
5.1.1 Polyp Segmentation
The public polyp segmentation benchmarks, including EndoScene (Vázquez et al., 2017), ETIS-Larib (Silva et al., 2014), Kvasir (Jha et al., 2021), and our private wireless capsule endoscope (WCE) datasets are adopted for cross imaging device generalization. As in Fig. 3, domain discrepancies among different datasets are evident.
EndoScene dataset (Vázquez et al., 2017) consists of 912 colonoscopy images collected by the devices of Olympus Q160AL and Q165L, Extra II video processor. Each colonoscopy image is equipped with pixel-level polyp segmentation ground truth.
ETIS-Larib dataset (Silva et al., 2014) includes 196 frames captured by Pentax 90i series, EPKi 7000 video processor.
Kvasir dataset (Jha et al., 2021) is composed of 1000 images and their polyp segmentation labels, which are obtained from colonoscopy video sequences and collected using endoscopic equipment at Vestre Viken Health Trust.
WCE dataset comprises 541 images collected from Prince of Wales Hospital with Medtronics Pillcam wireless capsule endoscope. The ground truths of polyp regions are labeled by two experts.
5.1.2 Surgical Instrument Segmentation
Porcine procedure instrument type segmentation datasets recorded from da Vinci XI system, including Endovis17 (Allan et al., 2019) and Endovis18 (Allan et al., 2020), are leveraged for cross-surgical site generalization, as illustrated in Fig. 4.
Endovis17 dataset (Allan et al., 2019) contains 1800 images captured from the abdominal porcine procedure, and three instrument type classes, including forceps, needle driver and scissor, are compatible with Endovis18.
Endovis18 dataset (Allan et al., 2020) is a kidney procedure dataset. It consists of 2384 images, and each image is equipped with the corresponding instrument type ground truths.
5.1.3 Evaluation Metrics
The performances of our method in polyp and instrument segmentation tasks are assessed by two commonly utilized metrics, including the Dice similarity coefficient (Dice) and Jaccard index (Jac). For both evaluation metrics, a higher value indicates a better segmentation result.
5.2 Implementation Details
Our methods are implemented with the PyTorch library on GeForce RTX 2080Ti GPU. For the polyp segmentation task, we adopt DeepLabv3+ (Chen et al., 2018) framework with the encoder ResNet101 pre-trained on ImageNet as our segmentation baseline network. To enlarge the training dataset, we utilize on-the-fly data augmentation, such as random flip, rotation, crop and scale. The augmented patches are then resized to 256\(\times \)256 for training. The SGD optimizer is adopted with an initial learning rate of 0.001 for the pre-trained encoder and 0.01 for the rest of the trainable parameters of the network with random initialization. Polynomial learning rate scheduling is adopted with a power of 0.9. We choose a batch size of 8 and a maximum epoch of 200. Regarding the hyper-parameters in the proposed InfProto method, we utilize \(\lambda =~\)1e-3 in Eq. (4). In hierarchical domain-adaptive classifier, \(\lambda _{\varvec{\mu }}\) and \(\lambda _{\varvec{\Sigma }}\) that control the contributions of instance-level, mini-batch-level and dataset-level data distributions are set as 0.2 and 0.8, respectively.
As for the surgical instrument segmentation task, DeepLabv2 (Chen et al., 2017) segmentation network with the pre-trained encoder ResNet101 is leveraged as our segmentation baseline. To fit the InfProto framework, the classifier in DeepLabv2 is replaced with FC layer. Following (Liu et al., 2021a), training images are first augmented with random flip and then resized to 320\(\times \)256 for training. We adopt an initial learning rate of 0.005 for the pre-trained encoder and 0.05 for the rest of the trainable parameters of the segmentation model with random initialization. SGD optimizer and polynomial learning rate scheduling with the power of 0.9 are leveraged for optimization. We choose a batch size of 4 and a maximum epoch number of 100. Hyper-parameters \(\lambda \), \(\lambda _{\varvec{\mu }}\), and \(\lambda _{\varvec{\Sigma }}\) are set as 1e-3, 0.2 and 0.8, respectively.
5.3 Comparison to SOTA Methods
5.3.1 Results on Polyp Segmentation
To validate the effectiveness of the proposed InfProto, we compare it with the baseline Deeplabv3+ (Chen et al., 2018), state-of-the-art fully supervised polyp segmentation methods (Fan et al., 2020; Guo et al., 2021b, 2022; Zhang et al., 2020b) and single DG methods (Choi et al., 2021; Kundu et al., 2021; Pan et al., 2019; Zhang et al., 2020a) in image segmentation, as shown in Table 2. It is obvious that InfProto shows satisfactory generalization performance with an average Dice of 71.517% and Jac of 63.414% and exhibits a significant average improvement of 15.568%, 16.152% in Dice and Jac compared with the baseline model DeepLabv3+. Notably, the level of domain discrepancy between EndoScene dataset and WCE dataset is the highest, and thus, the baseline model shows unsatisfactory generalization performance with 24.533 Dice score. On the contrary, our method can still perform well with a Dice of 55.239% in such a dilemma, demonstrating the superiority of the proposed InfProto framework in dealing with domain generalization problem.
State-of-the-art fully supervised polyp segmentation methods (Fan et al., 2020; Guo et al., 2021b, 2022; Zhang et al., 2020b) improve the performance through designing sophisticated network architectures with incremental computational costs. In contrast, our InfProto framework preserves the network architecture of backbone, and promotes generalizability by incorporating domain-adaptive statistical information and narrowing down intra-domain discrepancy. As a result, our approach significantly outperforms other polyp segmentation methods (Fan et al., 2020; Zhang et al., 2020b; Guo et al., 2021b, 2022) with average increments of 8.664%, 6.248%, 5.266%, 7.179% in Jac score on three target domain polyp segmentation datasets.
We then compare our method with (Choi et al., 2021; Kundu et al., 2021; Pan et al., 2019; Zhang et al., 2020a), which are representative approaches in addressing the single DG problem. Different from SW (Pan et al., 2019) that adapts the normalization layer of model, the proposed method elasticizes the classifier for various domains by incorporating the statistics of input instance and shows superior performance with 4.548% increment in Dice score. RobustNet (Choi et al., 2021) utilizes covariance statistics to disentangle the domain-specific style and domain-invariant content, while our method incorporates variance statistics to enhance the discriminability of the classifier. Consequently, the proposed method performs favorably against (Choi et al., 2021) with an average increment of 4.438% Dice score among three target domain datasets. Moreover, the proposed InfProto making full utilization of intra-domain discrepancies demonstrates better generalization capability than (Kundu et al., 2021; Zhang et al., 2020a) that augment multiple novel domains with increments of 4.692%, 4.262% in Jac.
We also provide the standard deviation values computed across all test samples for each generalization scenario. It is worth noting that the observed standard deviation scores are not very small, and there are two primary reasons for this. Firstly, The inherent variability in polyp images for segmentation tasks, as discussed in Guo et al. (2022), leads to substantial performance variability across different test samples. Secondly, the problems of domain gap and the easily confused polyp and normal tissues make the segmentation of unseen target domain data challenging, leading to undetected polyp regions and very low scores in some images. Despite these challenges, our proposed method demonstrates superior segmentation results with relatively small standard deviation values, revealing the robustness and effectiveness of our approach.
Qualitative comparison of polyp segmentation results by different methods evaluated on ETIS-Larib, Kvasir, and WCE datasets. From left to right are a the target domain images, results of methods b baseline model (Chen et al., 2018), c PraNet (Fan et al., 2020), d ACSNet (Zhang et al., 2020b), e DW-HieraSeg (Guo et al., 2021b), f NEIP (Guo et al., 2022), g SW (Pan et al., 2019), h BigAug (Zhang et al., 2020a), i RobustNet (Choi et al., 2021), j Vendorside (Kundu et al., 2021), and k the proposed InfProto framework. The ground truth labels are shown in (l) column for reference
We calculate the hit rate and miss rate in EndoScene\(\rightarrow \)ETIS-Larib scenario. Specifically, for each image, if the Dice score is above the threshold \(\tau \), the segmentation is considered a ‘hit’ for the ground truth polyp region; otherwise, it is considered a ‘miss’. Accordingly, I have varied \(\tau \) in [0.1, 0.3, 0.5, 0.7] and the corresponding results are shown in Fig. 5. Our method demonstrates the best performance in hit and miss rates compared to baseline methods. These results indicate that our proposed method does indeed attempt to segment each polyp individually, rather than focusing solely on the largest one.
We further illustrate typical polyp segmentation examples from three unseen domain datasets, as shown in Fig. 6. It is evident that the proposed InfProto method surpasses state-of-the-art polyp segmentation methods (Fan et al., 2020; Guo et al., 2021b, 2022; Zhang et al., 2020b) as well as single DG methods (Choi et al., 2021; Kundu et al., 2021; Pan et al., 2019; Zhang et al., 2020a) remarkably in the qualitative perspective. We can observe that baseline models are vulnerable to domain shifts, leading to missed polyp detection or false positive results. In drastic domain shift case (i.e., WCE dataset, \(9^{th}\)-\(12^{th}\) rows in Fig. 6), most baseline models fail to predict the polyp regions. In contrast, the proposed InfProto still finds key components successfully and obtains more reasonable segmentation predictions.
Qualitative comparison of surgical instrument segmentation results in Endovis17 \(\rightarrow \) Endovis18 and Endovis18 \(\rightarrow \) Endovis17 scenarios. From left to right are a the target domain images, results of methods b baseline model (Chen et al., 2018), c RAUNet (Ni et al., 2019), d LWANet (Ni et al., 2020), e MICCAI21 (Pissas et al., 2021), f DMNet (Wang et al., 2021a), g SW (Pan et al., 2019), h BigAug (Zhang et al., 2020a), i RobustNet (Choi et al., 2021), j Vendorside (Kundu et al., 2021), and k the proposed InfProto framework. The ground truth labels are shown in (l) column for reference
5.3.2 Results on Surgical Instrument Segmentation
In surgical instrument segmentation task, we first compare our method with the baseline model DeepLabv2 (Chen et al., 2017). Table 3 shows the comparison generalization performance in both Endovis17 \(\rightarrow \) Endovis18 and Endovis18 \(\rightarrow \) Endovis17 scenarios. Specifically, our InfProto method exhibits a significant average improvement of 17.009%, 16.624% in Dice and Jac compared with the baseline model among three instrument types in Endovis17 \(\rightarrow \) Endovis18. Then we implement the state-of-the-art fully supervised instrument segmentation methods (Ni et al., 2019, 2020; Pissas et al., 2021; Wang et al., 2021a), and our approach shows superior generalizability in comparison to fully supervised algorithms (Ni et al., 2019, 2020; Pissas et al., 2021; Wang et al., 2021a) with significant increments (i.e., the p-values are all smaller than.05) of 9.057%, 11.297%, 5.217%, 18.071% Jac in Endovis17 \(\rightarrow \) Endovis18. In the Endovis18 \(\rightarrow \) Endovis17, even though the forceps class segmentation performances of these fully supervised methods are higher than ours, needle driver is easily misclassified as forceps, which leads to high false positive rate for forceps class and extremely unsatisfactory performance for needle driver class, as validated in Fig. 7 (b-e) columns. Moreover, we observe that the proposed InfProto method outperforms favorably against existing single DG methods with significant improvements. For instance, InfProto surpasses (Choi et al., 2021) with average increments of 10.269%, 4.215% Jac score in two separate scenarios, demonstrating the good generalization capability of the proposed InfProto framework. Figure 7 presents typical instrument type segmentation results for a quantitative comparison of these different methods.
5.4 Analytical Study
5.4.1 Ablation Study on Polyp Segmentation
To inspect the role of each proposed component in InfProto framework, we conduct extensive ablation experiments, and comparison results are listed in Table 4. Firstly, the segmentation model with FC layer as the classifier is purely trained on the source domain data to investigate the primary performance gain that comes from the proposed framework, and we regard this model as the ‘Baseline’ model (2nd row), which is exactly the DeepLabv3+ network. Table 4 demonstrates the performance improvements in comparison to the ‘Baseline’ model, by gradually adding each proposed loss function into the pipeline. It is observed that each component contributes to the overall performance of our framework. Finally, the proposed InfProto framework (\(10^{th}\) row) achieves 74.157% Dice and 65.175% Jac scores, outperforming the ‘Baseline’ model by a significant margin.
Effectiveness of \({\mathcal {L}}_{Inf}^{I}\). With infinite prototypes being sampled from instance-level data distribution, the formed domain-adaptive classifier incorporates intra-class variance and contains rich semantic information. As shown in Table 4, compared with the ‘Baseline’ model that merely utilizes FC layer as the classifier, the proposed instance-level domain-adaptive classifier with \({\mathcal {L}}_{Inf}^{I}\) (3rd row) can achieve an improvement of 2.806% Jac score, verifying the effectiveness of incorporating instance-level statistics in DG task. Moreover, the comparisons of 5th \(\rightarrow \) 3rd rows, 6th \(\rightarrow \) 4th rows, and 9th \(\rightarrow \) 8th rows also demonstrate its superiority.
Qualitative comparison of segmentation results in EndoScene \(\rightarrow \) ETIS-Larib scenario. From left to right are a the target domain images from ETIS-Larib dataset, results of methods b baseline model (Chen et al., 2018), c w/o \({\mathcal {L}}_{HieraCR}\), d w/ \({\mathcal {L}}_{HieraCR}\), and e the ground truth label. Note that the number indicates the Dice(%) of per segmentation result
Effectiveness of \({\mathcal {L}}_{Inf}^{E}\). The proposed instance-level infinite prototype exchange (ProtoEx) strategy swaps the distribution information of two instances, and the segmentation result derived from the classifier formed by the exchanged prototypes is regularized by \({\mathcal {L}}_{Inf}^{E}\). As shown in the 3rd row of Table 4, merely regularizing the segmentation outputs with \({\mathcal {L}}_{Inf}^{E}\) gives rise to the performance increment compared with \({\mathcal {L}}_{Inf}^{I}\) (3rd row), insinuating that the proposed ProtoEx strategy can explicitly narrow down distribution shifts within the source domain to encourage the domain-agnostic feature extraction.
Effectiveness of \({\mathcal {L}}_{Inf}^{B}\). We then perform the mini-batch-level domain-adaptive classifier and regularize the model with \({\mathcal {L}}_{Inf}^{B}\), as shown in 5th row of Table 4. Since mini-batch infinite prototypes represent the common knowledge shared among variant micro source domains, the mini-batch statistics are more robust than the instance one, and thus, \({\mathcal {L}}_{Inf}^{B}\) shows better performance compared with \({\mathcal {L}}_{Inf}^{I}\) (3rd row). Moreover, combining \({\mathcal {L}}_{Inf}^{I}\) with \({\mathcal {L}}_{Inf}^{B}\) (7th row) exhibits the superior performance than utilizing either \({\mathcal {L}}_{Inf}^{I}\) (3rd row) or \({\mathcal {L}}_{Inf}^{B}\) (5th row), demonstrating that they are complementary in the proposed hierarchical domain-adaptive classifier.
Effectiveness of \({\mathcal {L}}_{HieraCR}\). The consistency regularization (HieraCR) is proposed to constrain the similarity of outputs from the hierarchical domain-adaptive classifier. Additionally, introducing \({\mathcal {L}}_{HieraCR}\) (10th row) leads to a significant performance gain with improvements of 5.357% in Dice score compared with 9 row. The promising increment demonstrates that the proposed InfProto framework can get rid of domain-specific characterizations and concentrate on extracting domain-agnostic features. Moreover, the qualitative comparison of ‘w/o \({\mathcal {L}}_{HieraCR}\)’ and ‘w/ \({\mathcal {L}}_{HieraCR}\)’ is illustrated in Fig. 9. In ‘w/o \({\mathcal {L}}_{HieraCR}\)’ (Fig. 9c), the segmentation model may under-represent (2nd row) or over-segment (3rd row) polyp regions due to the domain shift problem. In contrast, with the incorporated \({\mathcal {L}}_{HieraCR}\) (Fig. 9d), the segmentation model can effectively narrow down the domain discrepancy to enhance the generalization ability. Furthermore, the proposed consistency regularization (HieraCR) is composed of \({\mathcal {L}}\left( P^{I}, P^{E}\right) \), \({\mathcal {L}}\left( P^{I}, P^{B}\right) \), and \({\mathcal {L}}\left( P^{E}, P^{B}\right) \), and we ablate each component in Table 5. It is observed that each consistency regularization leads to performance gain.
5.4.2 Visualization of Deep Features
In order to qualitatively validate the effectiveness of the proposed InfProto framework, the dimensionality reduction algorithm, t-SNE (Maaten & Hinton, 2008), is performed to scatter the source and target domain data distributions. The comparison of the ‘Baseline’ model and our method in feature visualization is illustrated in Fig. 8. An ideal outcome would show no domain shift at all, as exemplified by the polyp data distributions of our method in (c, d). However, the domain shift in Fig. 8, despite the proposed method, indicates that there are still challenges in achieving fully domain-agnostic features. The optimized FC layer of the ‘Baseline’ model only incorporates statistics estimated from the source data, resulting in a fixed decision boundary that is unsuitable for discriminating the target domain distribution during inference. On the contrary, the proposed hierarchical domain-adaptive classifier can adaptively tailor the decision boundary for the target domain in the inference stage, offering substantial improvement. Additionally, even though t-SNE is a non-linear transformation, it is important to note that while translation directions between class-conditional data distributions are inconsistent in the ‘Baseline’ model, InfProto demonstrates consistent and approximately parallel shifts relative to the decision boundary, which is a positive step toward domain-agnostic feature learning.
5.4.3 Hyperparameter \(\lambda \)
The hyperparameter \(\lambda \) in Eq. (4) essentially controls the range of sampling infinite prototypes. To study how \(\lambda \) affects the performance of InfProto framework, we conduct a sensitivity test, where \(\lambda \) is varied in the range of [0.0, 5.0]. As shown in Fig. 10, with \(\lambda \) varying from 0.0 to 5.0, the performance of our method increases first and then decreases. Although a lower \(\lambda \) (e.g. \(1e-5\)) can reduce the effect of the scaled covariance matrix and is detrimental to performance, the corresponding results are still satisfactory, as illustrated in Fig. 10. We conjecture the reason is that, in this case, the sampled infinite prototypes are reliable. On the contrary, a larger \(\lambda \) (e.g. \(\lambda =1\) or 5) results in degraded performance. This is because the large value of \(\lambda \) leads to a diverse prototype sampling and may harm the discriminative ability of classifier. Moreover, we can observe that the performance of the proposed InfProto framework is relatively stable in a wide range of [1e-4, 5e-3], where \(\lambda \) value is small. This is because, for medical image segmentation tasks (e.g., polyp segmentation), abnormal (e.g., polyp) and normal tissues share similar characteristics, leading to close feature spaces between two classes. Therefore, a small \(\lambda \) value is suggested to obtain reliable sampled infinite prototypes and preserve the discriminative ability of the classifier.
5.4.4 Hyperparameters \(\lambda _{\varvec{\mu }}\), \(\lambda _{\varvec{\Sigma }}\)
As present in Sect. 4.1, the domain-adaptive classifier is formed by infinite prototypes sampled from \({\mathcal {N}} (\varvec{\mu }^{I}_{j}, ~ \varvec{\Sigma }^{I}_{j} )={\mathcal {N}} \left( \lambda _{\varvec{\mu }} \varvec{\mu }^{i}_{y} + (1-\lambda _{\varvec{\mu }}) \varvec{\mu }^{d}_{y}, ~ \lambda _{\varvec{\Sigma }} \varvec{\Sigma }^{i}_{y} + (1-\lambda _{\varvec{\Sigma }}) \varvec{\Sigma }^{d}_{y} \right) \), \({\mathcal {N}} (\varvec{\mu }^{B}_{j}, \varvec{\Sigma }^{B}_{j})={\mathcal {N}} (\lambda _{\varvec{\mu }} \varvec{\mu }^{b}_{y} + (1-\lambda _{\varvec{\mu }}) \varvec{\mu }^{d}_{y}, ~ \lambda _{\varvec{\Sigma }} \varvec{\Sigma }^{b}_{y} +\) \((1-\lambda _{\varvec{\Sigma }}) \varvec{\Sigma }^{d}_{y})\), where \(\lambda _{\varvec{\mu }}\) and \(\lambda _{\varvec{\Sigma }}\) control the contributions of instance-level, mini-batch-level and dataset-level data distributions. Notice that \(\lambda _{\varvec{\mu }}=0\) and \(\lambda _{\varvec{\Sigma }}=0\) indicates only the dataset level data distribution is reserved. The larger \(\lambda _{\varvec{\mu }}\) and \(\lambda _{\varvec{\Sigma }}\) are, the less contribution of dataset level data distribution will make. Herein, we vary \(\lambda _{\varvec{\mu }}\) and \(\lambda _{\varvec{\Sigma }}\) in the range of [0.0, 1.0] separately, and the resulting performance is shown in Fig. 11. From the Dice curve, the best performance is achieved when \(\lambda _{\varvec{\mu }}=0.8\) and \(\lambda _{\varvec{\Sigma }}=0.2\). This result reveals that instance-level and mini-batch-level 1st-moment information benefit the domain-adaptive property of the classifier more. Moreover, dataset-level 2nd-moment information characters more robust class-conditional feature variances. Hence, properly choosing small \(\lambda _{\varvec{\mu }}\) value and large \(\lambda _{\varvec{\Sigma }}\) value can promote the domain-adaptive ability and robustness of the classifier.
5.4.5 Tightness of the Upper Bound
As present in Sect. 3, the proposed InfProto derives the upper bound of the expected CE loss \({\mathcal {L}}_{CE}^{\infty }\) and leverages it as the surrogate loss for model optimization. Hence, the upper bound \({\mathcal {L}}_{Inf}\) is required to be tight to ensure that \({\mathcal {L}}_{CE}^{\infty }\) is minimized. To verify the tightness of the upper bound \({\mathcal {L}}_{Inf}\), we empirically compute \({\mathcal {L}}_{Inf}\) and \({\mathcal {L}}_{CE}^{\infty }\) over the training epochs, and the comparison curves are shown in Fig. 12. Note that when deriving \({\mathcal {L}}_{CE}^{\infty }\), we set \(K=10\) for approximation due to the limited computational resources. We can observe that \({\mathcal {L}}_{Inf}\) gives a tight upper bound. Since our choice of \(K=10\) is indeed far from the ideal case of \(K=\infty \), the observed Jensen gap may not appear small relative to \({\mathcal {L}}_{CE}^{K=10}\). However, the main point we wish to emphasize is the consistent behavior observed between our upper bound loss (\({\mathcal {L}}_{Inf}\)) and the expected CE loss (\({\mathcal {L}}_{CE}^{\infty }\)) throughout the training process. This alignment is a significant finding that underscores the practical effectiveness of our approach in optimizing the expected CE loss. With the minimization of \({\mathcal {L}}_{Inf}\) in the training process, the value of \({\mathcal {L}}_{CE}^{\infty }\) is optimized to converge simultaneously.
5.4.6 InfProto with Different Data Augmentation Methods
The proposed InfProto method represents a brand-new paradigm for single DG, complementary to the widely explored data/domain augmentation methods. To demonstrate this property, we incorporate four kinds of data augmentation methods to InfProto, including sharpening, brightness, contrast, and asymmetric mixup (Li et al., 2020). Notably, the former three methods stand as the best-performing augmentations in BigAug (Zhang et al., 2020a). As shown in Table 6, introducing data augmentation algorithms further boosts the performance of InfProto with increments of 0.896%, 0.467%, 1.409%, 0.828% Jac, verifying the complementary property.
5.4.7 Clinical and Practical Value of InfProto
The proposed single DG method, InfProto, aims to deal with the domain shift problem and release the heavy burden from annotating target domain data. Herein, we evaluate the amount of annotated target data required to reach a specific segmentation performance. Specifically, in EndoScene\(\rightarrow \)WCE scenario, InfProto and baseline models (Chen et al., 2017; Choi et al., 2021; Fan et al., 2020; Guo et al.,2021b; 2022; Kundu et al., 2021; Pan et al.,2019; Zhang et al., 2020a; b) optimized on EndoScene dataset are further fine-tuned with different numbers of labeled WCE images ranged in \(\{0, 10, 50, 100, 150, 200\}\). As illustrated in Fig. 13, it is obvious that the curve of our method is superior to other competed methods, verifying the generalizability of InfProto. Moreover, our approach requires the least number of labeled WCE images to achieve the same segmentation performance, giving a huge advantage over the baselines. For example, according to the horizontal green dotted line, InfProto requires 150 fewer target images than the baseline (Chen et al., 2017), and hence saves much more human effort in pixel-level annotation, validating the clinical value of InfProto. Furthermore, under the fully supervised learning (i.e., training with 405 labeled WCE images), our InfProto surpasses NEIP (Guo et al., 2022) with an increment of 3.004% in Jac, which reveals that the InfProto can still boost the performance even though optimized with sufficiently labeled target data. This reality demonstrates the explicitly extracted domain-agnostic features by InfProto exclude the redundant representations and concentrate on the semantically discriminative representations.
5.4.8 In-distribution Analysis
We further evaluate the performance of different methods when training and testing on the same WCE polyp dataset, as shown in ‘Fully supervised’ setting of the following Table 7. Note that ‘Single DG’ indicates the models are trained on EndoScene and tested on WCE polyp dataset, the performance of which is also provided in Table 7 for reference. We have two interesting observations from the comparison results. Firstly, InfProto performs best in the ‘Fully supervised’ setting. This is because our method minimizes distribution shifts within the limited WCE training data to extract domain-agnostic features, avoiding spurious features and alleviating the overfitting problem. Secondly, as for ‘Baseline’ method, the domain gap (i.e., between EndoScene and WCE datasets) leads the performance to drop from 36.554 to 16.205%, with a decrease of 20.349% in Jac score. The decrease of our proposed method is only 14.630%, demonstrating better cross-domain generalizability when only source domain data is available.
5.4.9 Comparison with Different Backbone Networks
We integrate the proposed InfProto into other advanced segmentation baselines: DeepLabv2 (Chen et al., 2017), MSNet (Zhao et al., 2021), CCBANet (Nguyen et al., 2021), and nnU-Net (Isensee et al., 2021) in polyp segmentation. Table 8 summarizes the segmentation comparison results (i.e., Dice and Jac), the practical time consumption [i.e., training time (h)], and the computational overhead [i.e., FLOPs (G)] of InfProto framework upon different backbones. The results are recorded on one GeForce RTX 2080 Ti GPU. It is observed that InfProto significantly improves the segmentation performance of Chen et al. (2017), Zhao et al. (2021), Nguyen et al. (2021), Isensee et al. (2021) with increments of 8.091%, 5.481%, 5.609%, 7.746% in Dice, respectively. These promotions validate that the proposed InfProto is a general method that could boost the performance of existing segmentation models. Moreover, it is obvious that InfProto boosts the performance upon the backbone ‘DeepLabv3+’ with an increment of 10.262% in Jac, while requiring only 0.60h additional training time and 0.91%G extra FLOPs. In addition, we can observe that InfProto involves more computational consumption on the backbone DeepLabv3+ compared to MSNet, CCBANet, and nnUNet. That is because the dimension of extracted features is relatively large in DeepLabv3+, and consequently, the corresponding size of estimated 1st-moment (\(\varvec{\mu }\)) and 2nd-moment (\(\varvec{\Sigma }\)) are large.
6 Limitations and Future Works
Firstly, the proposed ProtoEx, IBProto, and HieraCR strategies are based on the assumption that domain-agnostic features can be theoretically distilled from a single source domain exhibiting distribution shifts. When the source domain data is highly homogeneous, the improvement of the model may be limited. Moreover, we have demonstrated that the proposed InfProto method complements the data/domain augmentation methods. Therefore, our future works will involve domain randomization (Huang et al., 2021) or introduce extra knowledge, such as exemplars to guide domain augmentation. With the diversified source domain, the proposed strategies can be further boosted to mine the domain-agnostic features.
Secondly, our current research primarily focuses on addressing single domain generalization problems in the context of source and target domains with predefined categories. We acknowledge that in real-world medical scenarios, open-class and long-tail challenges are prevalent, where new, unforeseen categories may emerge, and some categories may have limited data. Future work should explore our InfProto to handle these challenges effectively, such as introducing unseen class semantic information for InfProto to build the semantic-adaptive classifier.
7 Conclusion
Conventional image segmentation models learned on existing data usually fail to generalize well on unseen domains. In this paper, we consider this practical yet challenging problem, i.e., single DG, and propose an adaptive infinite prototypes (InfProto) scheme. InfProto harnesses the high-order statistics and infinitely samples instance-specific prototypes to form the classifier for discriminability and generalizability facilitation, which is optimized by minimizing the upper bound of expected CE loss derived with theoretical justification. Reasoning that domain-adaptive classifier and domain-agnostic feature learning are key components in addressing the single DG problem, we first incorporate InfProto to design a hierarchical domain-adaptive classifier, which elasticizes the segmentation model for varying domains. Then, to extract domain-agnostic features, we formulate each instance within the source domain as a micro source domain, and further propose ProtoEx, IBProto, HieraCR strategies. Extensive experiments and ablation studies on polyp and instrument segmentation benchmarks comprehensively demonstrate the impact of InfProto.
Notes
InfProto code: https://github.com/CityU-AIM-Group/InfProto.
References
Ahmed, S. M., Raychaudhuri, D. S., Paul, S., et al. (2021). Unsupervised multi-source domain adaptation without access to source data. In CVPR (pp. 10103–10112).
Allan, M., Shvets, A., Kurmann, T., et al. (2019). 2017 robotic instrument segmentation challenge. arXiv preprint arXiv:1902.06426.
Allan, M., Kondo, S., Bodenstedt, S., et al. (2020). 2018 robotic scene segmentation challenge. arXiv preprint arXiv:2001.11190.
Bai, H., Sun, R., Hong, L., et al. (2021). Decaug: Out-of-distribution generalization via decomposed feature representation and semantic augmentation. In AAAI (pp. 6705–6713).
Chang, Y. T., Wang, Q., Hung, W. C., et al. (2020). Weakly-supervised semantic segmentation via sub-category exploration. In CVPR (pp. 8991–9000).
Chen, L. C., Papandreou, G., Kokkinos, I., et al. (2017). Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Transactions on Pattern Analysis and Machine Intelligence, 40(4), 834–848.
Chen, L. C., Zhu, Y., Papandreou, G., et al. (2018). Encoder–decoder with atrous separable convolution for semantic image segmentation. In ECCV (pp. 801–818).
Chen, Y., & Xia, Y. (2021). Iterative sparse and deep learning for accurate diagnosis of Alzheimer’s disease. Pattern Recognition, 116(107), 944.
Cheng, Y., Wei, F., Bao, J., et al. (2021). Dual path learning for domain adaptation of semantic segmentation. In ICCV (pp. 9082–9091).
Choi, S., Jung, S., Yun, H., et al. (2021). Robustnet: Improving domain generalization in urban-scene segmentation via instance selective whitening. In CVPR (pp. 11580–11590).
Creager, E., Jacobsen, J. H., Zemel, R. (2021). Environment inference for invariant learning. In ICML (pp. 2189–2200).
Dubey, A., Ramanathan, V., Pentland, A., et al. (2021). Adaptive methods for real-world domain generalization. In CVPR (pp. 14340–14349).
Fan, D. P., Ji, G. P., Zhou, T., et al. (2020). Pranet: Parallel reverse attention network for polyp segmentation. In MICCAI (pp. 263–273).
Fan, X., Wang, Q., Ke, J., et al. (2021). Adversarially adaptive normalization for single domain generalization. In CVPR (pp. 8208–8217).
Fourure, D., Emonet, R., Fromont, E., et al. (2017). Residual conv-deconv grid network for semantic segmentation. In BMVC.
Guo, X., Liu, J., & Yuan, Y. (2021). Semantic-oriented labeled-to-unlabeled distribution translation for image segmentation. IEEE Transactions on Medical Imaging, 41(2), 434–445.
Guo, X., Yang, C., & Yuan, Y. (2021). Dynamic-weighting hierarchical segmentation network for medical images. Medical Image Analysis, 73(102), 196.
Guo, X., Chen, Z., Liu, J., et al. (2022). Non-equivalent images and pixels: Confidence-aware resampling with meta-learning mixup for polyp segmentation. Medical Image Analysis, 78(102), 394.
Han, J., Luo, P., Wang, X. (2019). Deep self-learning from noisy labels. In ICCV (pp. 5138–5147).
He, J., Jia, X., Chen, S., et al. (2021). Multi-source domain adaptation with collaborative learning for semantic segmentation. In CVPR (pp. 11008–11017).
Huang, J., Guan, D., Xiao, A., et al. (2021). Fsdr: Frequency space domain randomization for domain generalization. In CVPR (pp. 6891–6902).
Isensee, F., Jaeger, P. F., Kohl, S. A., et al. (2021). nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nature Methods, 18(2), 203–211.
Jha, D., Smedsrud, P. H., Johansen, D., et al. (2021). A comprehensive study on colorectal polyp segmentation with resunet++, conditional random field and test-time augmentation. IEEE Journal of Biomedical and Health Informatics, 25(6), 2029–2040.
Kang, G., Jiang, L., Wei, Y., Yang, Y., & Hauptmann, A. (2020). Contrastive adaptation network for single-and multi-source domain adaptation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 44(4), 1793–1804.
Kim, M., Byun, H. (2020). Learning texture invariant representation for domain adaptation of semantic segmentation. In CVPR (pp. 12975–12984).
Kundu, J.N., Kulkarni, A., Singh, A., et al. (2021). Generalize then adapt: Source-free domain adaptive semantic segmentation. In ICCV (pp. 7046–7056).
Li, G., Jampani, V., Sevilla-Lara, L., et al. (2021). Adaptive prototype learning and allocation for few-shot segmentation. In CVPR (pp. 8334–8343).
Li, L., Gao, K., Cao, J., et al. (2021). Progressive domain expansion network for single domain generalization. In CVPR (pp. 224–233).
Li, S., Xie, M., Gong, K., et al. (2021). Transferable semantic augmentation for domain adaptation. In CVPR (pp. 11516–11525).
Li, X., Zhong, Z., Wu, J., et al. (2019). Expectation-maximization attention networks for semantic segmentation. In ICCV (pp. 9167–9176).
Li, Z., Kamnitsas, K., & Glocker, B. (2020). Analyzing overfitting under class imbalance in neural networks for image segmentation. IEEE Transactions on Medical Imaging, 40(3), 1065–1077.
Li, Z., Sun, Y., Zhang, L., et al. (2021). CTNet: Context-based tandem network for semantic segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 44(12), 9904–9917.
Liu, J., Guo, X., & Yuan, Y. (2021). Graph-based surgical instrument adaptive segmentation via domain-common knowledge. IEEE Transactions on Medical Imaging, 41(3), 715–726.
Liu, Y., Zhang, X., Zhang, S., et al. (2020). Part-aware prototype network for few-shot semantic segmentation. In ECCV (pp. 142–158).
Liu, Y., Zhang, W., & Wang, J. (2021). Source-free domain adaptation for semantic segmentation. In CVPR (pp. 1215–1224).
Maaten, Lvd, & Hinton, G. (2008). Visualizing data using t-SNE. Journal of Machine Learning Research, 9, 2579–2605.
Minaee, S., Boykov, Y. Y., Porikli, F., Plaza, A., Kehtarnavaz, N., & Terzopoulos, D. (2021). Image segmentation using deep learning: A survey. IEEE Transactions on Pattern Analysis and Machine Intelligence, 44(7), 3523–3542.
Nguyen, T. C., Nguyen, T. P., Diep, G. H., et al. (2021). Ccbanet: Cascading context and balancing attention for polyp segmentation. In MICCAI (pp. 633–643).
Ni, Z. L., Bian, G. B., Zhou, X. H., et al.(2019). Raunet: Residual attention u-net for semantic segmentation of cataract surgical instruments. In ICONIP (pp. 139–149).
Ni, Z. L., Bian, G. B., Hou, Z. G., et al. (2020). Attention-guided lightweight network for real-time segmentation of robotic surgical instruments. In ICRA (pp. 9939–9945).
Pan, X., Zhan, X., Shi, J., et al. (2019). Switchable whitening for deep representation learning. In ICCV (pp. 1863–1871).
Pandey, P., Raman, M., Varambally, S., et al. (2021). Generalization on unseen domains via inference-time label-preserving target projections. In CVPR (pp. 12924–12933).
Pissas, T., Ravasio, C. S., Cruz, L. D., et al. (2021). Effective semantic segmentation in cataract surgery: What matters most? In MICCAI (pp. 509–518).
Qiao, F., Zhao, L., & Peng, X. (2020). Learning to learn single domain generalization. In CVPR (pp. 12556–12565).
Ronneberger, O., Fischer, P., & Brox, T. (2015). U-net: Convolutional networks for biomedical image segmentation. In MICCAI (pp. 234–241).
Sagar, A., & Soundrapandiyan, R. (2021). Semantic segmentation with multi scale spatial attention for self driving cars. In ICCV (pp. 2650–2656).
Silva, J., Histace, A., Romain, O., et al. (2014). Toward embedded detection of polyps in wce images for early diagnosis of colorectal cancer. International Journal of Computer Assisted Radiology and Surgery, 9(2), 283–293.
Sohoni, N., Dunnmon, J., Angus, G., et al. (2020). No subclass left behind: Fine-grained robustness in coarse-grained classification problems. NeurIPS, 33, 19339–19352.
Su, Z., Yao, K., Yang, X., et al. (2023). Rethinking data augmentation for single-source domain generalization in medical image segmentation. In AAAI (pp. 2366–2374).
Truong, T. D., Le, N., Raj, B., et al. (2023). Fredom: Fairness domain adaptation approach to semantic scene understanding. In CVPR (pp. 19988–19997).
Vázquez, D., Bernal, J., Sánchez, F. J., et al. (2017). A benchmark for endoluminal scene segmentation of colonoscopy images. Journal of Healthcare Engineering, 2017, 4037190.
Volpi, R., Namkoong, H., Sener, O., et al. (2018). Generalizing to unseen domains via adversarial data augmentation. In: NeurIPS, pp 5339–5349.
Wang, J., Sun, K., Cheng, T., Jiang, B., Deng, C., Zhao, Y., Liu, D., Mu, Y., Tan, M., & Wang, X. (2020). Deep high-resolution representation learning for visual recognition. IEEE Transactions on Pattern Analysis and Machine Intelligence, 43(10), 3349–3364.
Wang, J., Jin, Y., Wang, L., et al. (2021). Efficient global-local memory for real-time instrument segmentation of robotic surgical video. In MICCAI (pp. 341–351).
Wang, W., Yin, B., Yao, T., et al. (2021). Delving into data: Effectively substitute training for black-box attack. In Proceedings of the CVPR (pp. 4761–4770).
Wang, Y., Huang, G., Song, S., Pan, X., Xia, Y., & Wu, C. (2021). Regularizing deep networks with semantic data augmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 44(7), 3733–3748.
Xu, Q., Zhang, R., Zhang, Y., et al. (2021). A fourier-based framework for domain generalization. In CVPR (pp. 14383–14392).
Xu, W., Xian, Y., Wang, J., et al. (2020). Attribute prototype network for zero-shot learning. In NeurIPS (pp. 21969–21980).
Yang, B., Liu, C., Li, B., et al. (2020). Prototype mixture models for few-shot semantic segmentation. In ECCV.
Zhang, L., Wang, X., Yang, D., et al. (2020). Generalizing deep learning for medical image segmentation to unseen domains via deep stacked transformation. IEEE Transactions on Medical Imaging, 39(7), 2531–2540.
Zhang, R., Li, G., Li, Z., et al. (2020). Adaptive context selection for polyp segmentation. In MICCAI (pp. 253–262).
Zhang, Y., David, P., Foroosh, H., et al. (2019). A curriculum domain adaptation approach to the semantic segmentation of urban scenes. IEEE Transactions on Pattern Analysis and Machine Intelligence, 42(8), 1823–1841.
Zhao, H., Shi, J., Qi, X., et al. (2017). Pyramid scene parsing network. In CVPR (pp. 2881–2890).
Zhao, L., Liu. T., Peng, X., et al. (2020). Maximum-entropy adversarial data augmentation for improved generalization and robustness. In NeurIPS.
Zhao, L., Peng, X., Chen, Y., et al. (2020). Knowledge as priors: Cross-modal knowledge generalization for datasets without superior knowledge. In CVPR (pp. 6528–6537).
Zhao, X., Zhang, L., & Lu, H. (2021). Automatic polyp segmentation via multi-scale subtraction network. In MICCAI (pp. 120–130).
Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., et al. (2019). Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Transactions on Medical Imaging, 39(6), 1856–1867.
Acknowledgements
This work was supported by Innovation and Technology Commission- Innovation and Technology Fund ITS/229/22 and Hong Kong Research Grants Council (RGC) General Research Fund 11211221 14204321. We thank the editor and reviewers for their valuable feedback and suggestions, which greatly improved this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Ehsan Adeli.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guo, X., Liu, J. & Yuan, Y. Infproto-Powered Adaptive Classifier and Agnostic Feature Learning for Single Domain Generalization in Medical Images. Int J Comput Vis (2024). https://doi.org/10.1007/s11263-024-02158-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11263-024-02158-9