Abstract
While huge strides have recently been made in language-based machine learning, the ability of artificial systems to comprehend the sequences that comprise animal behavior has been lagging behind. In contrast, humans instinctively recognize behaviors by finding similarities in behavioral sequences. Here, we develop an unsupervised behavior-mapping framework, SUBTLE (spectrogram-UMAP-based temporal-link embedding), to capture comparable behavioral repertoires from 3D action skeletons. To find the best embedding method, we devise a temporal proximity index (TPI) as a new metric to gauge temporal representation in the behavioral embedding space. The method achieves the best TPI score compared to current embedding strategies. Its spectrogram-based UMAP clustering not only identifies subtle inter-group differences but also matches human-annotated labels. SUBTLE framework automates the tasks of both identifying behavioral repertoires like walking, grooming, standing, and rearing, and profiling individual behavior signatures like subtle inter-group differences by age. SUBTLE highlights the importance of temporal representation in the behavioral embedding space for human-like behavioral categorization.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Behavior categorization is the process of grouping different behavioral patterns into meaningful categories based on their characteristics (Pereira et al., 2020; Shima et al., 2007). Such categorization is a key learning attribute of the human brain and involves identifying common features that are shared across different behaviors while also creating meaningful distinctions between behavioral categories. This process is essential to how we understand and interact with the environment around us. Humans can categorize complex behaviors intuitively by observing only a few coherently moving key points (Johansson, 1973). In contrast, it is far more difficult for machines to categorize the complex behavior of moving humans and animals, despite recent advances that enable machines to recognize 3D points accurately (Dunn, 2021; Huang, 2021; Kim et al., 2022; Marks, 2022; Marshall, 2021; Nath, 2019). Since we have yet to understand how humans intuitively recognize behavioral similarities, enabling machines to understand natural behavior by modeling human learning remains a challenge.
Recent studies have tried to establish machine-learning frameworks for categorizing behavior (Bohnslav, 2021; Hsu & Yttri, 2021; Luxem, 2022; Segalin et al., 2021), particularly with supervised behavior mapping that uses human-annotated labels (Bohnslav, 2021; Segalin et al., 2021). However, this supervised approach is labor-intensive and prone to observer bias. Indeed, the process of manually annotating behavioral patterns can be time-consuming (minimum 3–4x the video’s duration varying performance across days), especially for large datasets, and the results may be influenced by the subjective judgments of the annotators (Segalin et al., 2021; Sturman, 2020). To overcome these limitations, there has been a growing interest in exploring unsupervised learning methods (Berman et al., 2014; Brattoli, 2021; Hsu & Yttri, 2021; Nilsson et al., 2020; Segalin et al., 2021; Wiltschko, 2015; Weinreb et al., 2023). These methods, which do not require extensive human annotation, offer a promising alternative by potentially reducing labor and mitigating observer bias, thus representing a significant direction for future research in this field.
1.1 Need of Temporal Representation in Behavioral Embedding Spaces
Without carefully designed objectives or constraints, unsupervised methods cannot guarantee desired outcomes, as they cannot directly access labeled target outputs. Instead, the incorporation of domain knowledge or additional constraints is required to guide unsupervised algorithms toward a more meaningful solution. In most cases, such methods aim to understand the underlying structure of the data to extract meaningful patterns and relationships. This is achieved through feature extraction, dimensionality reduction, and clustering.
Such data representation requires a robust feature embedding space to ensure the reliability and validity of the results. An embedding space allows relevant features and relationships between entities to be represented so that similar entities are close together and dissimilar entities are far apart (De Oliveira & Levkowitz, 2003). As behavior is a complex and dynamic process that changes over time, it is challenging to quantify its temporal structure without a clear ground truth (Pereira et al., 2020). Therefore, unsupervised behavior mapping may require effective temporal representation.
1.2 Lacking Consensus on Forming Behavioral Embedding Space
While numerous approaches have aimed to construct an embedding space for behaviors, Berman et al. made a significant contribution by generating maps of fly behavior based on an unsupervised approach (Berman et al., 2016, 2014). They successfully assigned behavioral features to a low-dimensional embedding space using wavelet spectrograms for feature engineering and t-stochastic neighbor embedding (t-SNE) for nonlinear dimension reduction (Berman et al., 2014). From this embedding space, they employed predictive information bottleneck strategies to identify biologically meaningful repertoires (Berman et al., 2016).
While Berman et al. significantly influenced later studies and led to the emergence of other variants (Bala, 2020; Berman et al., 2016; Cande, 2018; Dunn, 2021; Günel, 2019; Hsu & Yttri, 2021; Klaus, 2017; Luxem, 2022; Marshall, 2021; Pereira, 2019; Zimmermann et al., 2020), the field has not defined criteria or reached a consensus on what constitutes a good behavioral embedding space (Table 1). Importantly, how the embedding space is designed is critical for predicting meaningful and reliable categories. In the absence of a standard evaluation method, however, previous research has yet to evaluate the quality of embedding spaces and has instead focused on optimizing the downstream task.
Despite the increasing use of UMAP (McInnes et al., 2018; York et al., 2020; Karashchuk et al., 2021; DeAngelis et al., 2019; Bala, 2020; Hsu & Yttri, 2021; Huang, 2021), which can capture more of the global structure in data compared to t-SNE, its effectiveness in behavioral science has not been thoroughly examined. While there have been some studies comparing these two methods (Hsu & Yttri, 2021; Luxem, 2022), a more comprehensive analysis combined with various feature extraction methods, specifically in the context of behavioral science is still warranted.
Evaluation strategy for temporal representation in the behavior embedding spaceA Conceptual illustration of an example behavior sequence for the 3D action skeleton trajectory of a mouse, where each color represents stereotyped behavioral repertoires. B Illustration of example pattern trajectories in the behavior embedding space, indicating embedding quality can determine more efficient trajectories; left, good embedding space with each cluster containing the same behavioral states with efficient temporal trajectories; right, bad embedding space with each cluster contains the different behavioral states with inefficient temporal trajectories. C Illustration of example temporal connectivity in the behavior embedding space represented with transition probabilities from source to target clusters; left, good temporal connectivity with closer distances between clusters as the transition probability increases; right, bad temporal connectivity with greater distances between clusters as the transition probability increases. D Temporal Proximity Index (TPI), for assessing the temporal connectivity of the behavior embedding space. E Schematic workflow for unsupervised behavior categorization and evaluation in this study
1.3 Humans Categorize Behavior Using Temporal Information
Humans can learn new concepts from just a few samples by leveraging prior knowledge and using probabilistic reasoning to build new concepts from the observed examples. This process is thought to involve the utilization of temporal information and transition probabilities for sequence coding, which allows us to represent and utilize the probabilities of future events and the time intervals between those events to guide our choices and behavior. The human brain constructs categorical representations, potentially serving as higher-level feature embeddings (Chang, 2010; Shima et al., 2007; Zhang et al., 2020), and tracks the transition probability between events to anticipate future events, exhibiting a stronger response to unexpected events compared to expected events.
Introducing human-based constraints into machine-learning models may improve performance, as research has shown that machines can achieve human-level concept learning through probabilistic program induction (Lake et al., 2015) and natural language description (Kojima et al., 2022; Zhou et al., 2022). The process by which humans categorize behaviors is not fully understood, but it is known that the brain uses temporal dynamic information encoded in embedding space for categorization. For example, a temporal choice task has been utilized to investigate temporal and probabilistic calculations in both mice and humans (Dehaene et al., 2015). As we further investigate the human brain’s ability to process and utilize temporal information and transition probabilities (Dehaene et al., 2015), we can potentially enhance machine learning models to mimic human learning capabilities, bridging the gap between human and machine cognition.
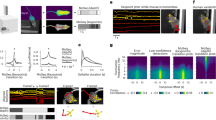
SUBTLE method gives the highest temporal connectivity score in embedding space A Schematic flowchart for preparing various feature embedding methods. (1) 3D action skeletons were extracted with the AVATAR recording system. (2) Kinematic features and wavelet spectrograms were extracted from raw coordinates. (3) Nonlinear mapping with t-SNE and UMAP algorithms. B Inter-cluster transition probabilities are visualized on embedding spaces as the number of clusters of k-Means clustering increases. Arrows with thickness representing state transition probabilities (Colors represent different clusters). C TPI scores were calculated with three input features (coordinates, kinematics, spectrogram) and two nonlinear embedding methods (t-SNE, UMAP; n = 8, number of runs). The inset shows the magnified plot of the black box containing t-SNE results. D Comparison of the second largest eigenvalue \(\lambda _2\) and its characteristic time \(t_2\) of the transition matrix obtained from spectrogram-UMAP and spectrogram-t-SNE with respect to increasing cluster numbers in k-Means clustering (n = 10, number of animals). Welch’s t-test, ***p < 0.001. E Effect of increasing perplexity in t-SNE algorithm (n = 8). Inset, the magnified plot from \(\log _2k\) = 4, 5, 6, 7 (n = 8, number of runs). F Effect of increasing neighbor in UMAP algorithm (n = 8, number of runs). G Comparison of computational efficiency between spectrogram-UMAP and spectrogram-t-SNE with respect to Neighbors or Perplexity hyperparameters (15, 30, 50, 100). Values were normalized by UMAP with neighbors 15. Data represent mean ± SD. H Effect of increasing min_dist parameter in UMAP algorithm (n = 8, number of runs)
SUBTLE effectively reflects kinematic feature representation in embedding space A Schematic illustration of kinematic similarity analysis in the behavioral state embedding space. Note that kinematic features were not directly included while constructing the embedding space. B The relationship between inter-embedding kinematic feature similarity \(\cos _\alpha \) and normalized embedding distance \(\Delta {x}\) (n = 10, number of animals). Note that data were calculated by pair-wise distances and cosine similarities for all data points per animals. C Perspective contrast of inter-embedding kinematic similarity in mice (n = 10, number of animals). Welch’s t-test, **p < 0.01. D The first principal component of kinematic features was plotted on the embedding spaces constructed from spectrogram-t-SNE and spectrogram-UMAP. Data represent mean ± SD
In summary, the contributions of this paper can be summarized as follows:
-
Introduction of a TPI as a novel metric for evaluating temporal representation in behavioral embedding spaces.
-
Demonstrating the superiority of UMAP-spectrogram over t-SNE-spectrogram in capturing kinematic and temporal structures within behavioral data.
-
Proposes SUBTLE method that emphasizes the significance of temporal representation for behavior categorization akin to humans.
-
Utilizes SUBTLE to derive meaningful biological insights from 3D action skeletons of young and adult animals.
-
Development of a user-friendly website with GUI functionalities for better visualization of animal behavior.
SUBTLE-based subclusters effectively capture biologically meaningful features A State transition map of Phenograph subclusters in embeddings of spectrogram-t-SNE and spectrogram-UMAP. Arrows with thickness represents state transition probabilities. B State retention map of Phenograph subclusters in embeddings of spectrogram-t-SNE and spectrogram-UMAP. Intensity represents state retention rates. C Human-annotated behavior categories in spectrogram-t-SNE and spectrogram-UMAP. Colors represent different categorical annotations. D–E Dendrogram of inter-subject behavior transition similarity measured by Frobenius norm of two different transition probability matrices \(P_A\) and \(P_B\). Asterisks with the same colors denote the same subject with different recording periods, young and adult. Heights represent the similarity distance measured by Frobenius matrix norm. F Comparison of minimum spanning nodes to join the other class in the dendrogram (n = 19 and 19). Welch’s t-test, *p < 0.05. G Empirical cumulative distribution function of human-annotated behavioral states against retention rate. Data represent mean ± SD
Comparison of SUBTLE-based superclusters and human-annotated behavior categories A Hierarchical clustering with deterministic information bottleneck method, using lag \(\tau = 2{t_c} \approx 20\). Identified representative superclusters (n = 2, 4, 6) in UMAP. B Human-annotated behavioral categories in UMAP. C Example transition probability matrix of subclusters grouped by 6 superclusters (n = 10, number of animals). D Sankey diagram flowing horizontally from left to right with the increasing number of clusters. E Example ethogram showing 30 s of mouse behavior. F Comparison of superclusters with human-annotated behavioral categories using adjusted rand index (top) and normalized mutual information (bottom) (n = 8, number of samples). G Differentially exhibited kinematic features of young (n = 8) and adult (n = 11) mice. Welch’s t-test, *p < 0.05, **p < 0.01, ***p < 0.001. V, node(joint) properties; E, edge(bone) properties; A, angle properties; superscript denotes n-th temporal derivatives; subscript denotes node indices; see method for details. H The proportion of significantly different features in each behavioral cluster. Data represent mean ± SD
Importance of temporal sequence in human-like behavior categorization. A Conceptual illustration of shuffling the behavioral states with fixed chunk size (top) and shuffled temporal connectivity in behavioral embedding space (bottom). B Evaluation of temporal representation with TPI metric in shuffled embedding space with varying numbers of chunk sizes (n = 8). C Evaluation of clustering quality of superclusters using the shuffled embedding with varying numbers of chunk sizes (n = 8). Data represent mean ± SD
2 Results
2.1 Evaluating Temporal Proximity of Behavioral Embeddings
To enable machines to categorize behaviors in a manner similar to that of humans, we have developed a simple metric to measure temporal representation in behavioral embedding space. To evaluate the quality of the embedding space, we assume that a good embedding space should preserve the temporal representation of behaviors, which is reflected in the distances between embeddings (Fig. 1A, B). Based on this premise, we have developed an evaluation method to quantify the temporal connectivity (Fig. 1C, D) by calculating the transition probability between two clusters in the space. We consider two cluster centers to be temporally linked in the embedding space if they are in close proximity and have a high transition probability. Given the number of clusters (n) and their centroids \(\{c_1,\cdots , c_{n}\}\), we define the TPI for quantifying temporal representation as the following equation:
\(p_{ij}\) is a transition probability from source cluster \(c_i\) to target cluster \(c_j\). \(w_{ij}\) is a proximity measure, which is defined as
\(w_{ij}\) is calculated from source to target cluster distances \(\textbf{d}_i = \{ d_{ij, j \ne i} \mid j=1, \cdots , N\}\) where \(d_{ij}=\Vert c_i-c_j\Vert _2\), an \(L_2\)-norm or euclidean distance between two clusters. Thus, \(w_{ij}\) represents the relative proximity between the source cluster i and the target cluster j, measured by Softmax of the inverse distance \(1/d_{ij}\). Overall, \(\text {TPI}\) can be interpreted as the sum of proximity-weighted transition probabilities. To implement the concept of TPI, we designed a workflow similar to the previously described unsupervised behavior mapping framework by creating feature embeddings, subclusters, and superclusters (Fig. 1D) (Berman et al., 2014, 2016).
Comparative Visualization of Dimensionality Reduction and Clustering Techniques in Behavioral Data. A Side-by-side TSNE-S and UMAP-S visualization of Fit3D dataset with labels. B, D, F Temporal connectivity analysis in various animal datasets including humans, rats, and mice (n = 6). C Comparison of clustering quality via ARI and NMI metrics (n = 6), in TSNE-S and UMAP-S in combination with k-Means clustering or Phenograph clustering. E, G UMAP scatter plots illustrating subclusters with Phenograph clustering and superclusters after DIB
2.2 Spectrogram-UMAP, A Powerful Temporal Embedding Method
To create feature embedding from action skeletons, we obtained 3D action skeleton trajectories from freely moving mice (\(N=10\), \(\approx \)120,000 video frames) using the AVATAR system (Kim et al., 2022) (Fig. 2A, left). The recording system (Fig. 2A, left) utilizes five cameras to record multi-view images, while AVATARnet captures key points via object detection and reconstructs their key point trajectories in 3D Euclidean space (Fig. S1). For the input features (Fig. 2A, right), we extracted additional features from (ii) Morlet wavelet spectrograms and (iii) kinematic features from (i) raw coordinates of 3D action skeleton trajectories (See Sect. 5.4.1 for spectrogram and Sect. 2.3.1 for kinematic feature generation). For the nonlinear mapping (Fig. 2A, right bottom), we applied (1) t-stochastic neighbor embedding (t-SNE) (Van der Maaten & Hinton, 2008) or (2) uniform manifold approximation and projection (UMAP) (McInnes et al., 2018) on the three input features to generate six combinations of feature embeddings. Two examples of feature embeddings are shown in Fig. 2B.
Next, we evaluated the temporal representation of each embedding space using TPI (Eq. 1). By conducting k-Means clustering with increasing cluster numbers, k, we could enhance the resolution of temporal connectivity (Fig. 2B). While the spectrogram-UMAP feature embedding showed a significant improvement in TPI scores at high k, spectrogram-t-SNE did not (Fig. 2B). Overall, the results demonstrated that using the wavelet spectrogram in combination with UMAP resulted in the highest TPI value or the best temporal representation, outperforming the other five combinations (Fig. 2C). These results indicate that the wavelet spectrogram best reflects the temporal representation compared to raw coordinates or kinematic features (Fig. 2C).
When it comes to the nonlinear mapping, we found that UMAP has higher TPI values than t-SNE (Fig. 2B, C), likely due to its ability to preserve the global structure (Kobak & Linderman, 2021). To test this possibility, we systematically altered the global structure of embedding space by changing the main hyperparameters for t-SNE (with perplexity) and UMAP (with neighbors). Regrettably, there is no universally optimal number of hyperparameters for neighbors in UMAP and perplexity in t-SNE, general recommendation ranges from 15 to 100 (common default values for neighbors = 15 and perplexity = 30). Generally, increasing the values of these hyperparameters will prioritize the preservation of global structure in the embedding space at the expense of local structure. While we observed a general increase in TPI scores for both t-SNE and UMAP by increasing these hyperparameters, the increase was more pronounced for UMAP (Fig. 2E, F), implying that the ability of UMAP to better preserve global structure compared to t-SNE contributes to enhancing temporal connectivity in the embedding space. In addition, UMAP was computationally efficient compared to t-SNE, highlighting its practical advantage (Fig. 2G). Balancing computational cost and TPI values, we set the UMAP neighbors parameter to 50, optimizing the preservation of global structure and efficiency in analyzing temporal connectivity. While decreasing the min_dist parameter in UMAP was effective in increasing TPI (Fig. 2H), we set the default value of min_dist as 0.1 throughout this work.
Next, to investigate the time scale of temporal dynamics in this embedding space, we measured the characteristic time (i.e., information decay of the behavioral state) of state transitions following the previous approach (Berman et al., 2016). We consider the number of clusters, k, to be behavioral states, and if the dynamics of the behavioral states follow Markovian dynamics, the transition from one state to another is independent of history. However, the Markov model has almost no information beyond 40 steps (2 s), while the transition matrices from actual data for \(\tau \) = 40 (2 s) and \(\tau \) = 400 (20 s) retain information (See Fig 12A). These results support the notion that the dynamics of freely moving mice are not Markovian, and they exhibit a much longer time (at least more than 2 s) structure than the Markovian model. In the Markovian system, the transition matrix \(\textbf{T}\) provides a complete description of the dynamics. The slowest time scale in the Markovian system is determined by the second largest eigenvalue, \(|\lambda _2 |\), which results in a characteristic decay time (the average time point at which 1/e of information is lost), \( t_c = -1 / \log {|\lambda _2 |}\) (Berman et al., 2016). We obtained the \(|\lambda _2 |\), and \(t_c\) from \(\textbf{T}\) constructed from k number of clusters for spectrogram-UMAP and spectrogram-t-SNE. We found that spectrogram-UMAP gives a higher characteristic time than the spectrogram-t-SNE at \(\log _2{k>4}\) (Fig. 2D). This result implies that information on behavioral states decays significantly more slowly in spectrogram-UMAP than in spectrogram-t-SNE. Based on these results, we conclude that the spectrogram-UMAP-based temporal-link embedding (SUBTLE) offers better preservation of temporal representation in the behavioral embedding space.
2.3 Evaluating Kinematic Similarity of Behavioral Embeddings
As kinematic features provide a quantitative description of how an organism moves, we hypothesized that a good behavioral embedding space should manifest the kinematic characteristics. Yet, the definition of kinematic features varies among researchers in accumulated studies, and these uniquely hand-crafted features complicate objective comparisons and hinder the reproducibility of research outcomes. To resolve this issue, we have defined kinematic features in a systematic way while maintaining high explainability.
2.3.1 Defining Kinematic Features
Here we provide a general definition of kinematic features. A mouse action skeleton dataset is given as continuous frames of multiple joint \(v_t\) of 2D \((x_t, y_t)\) or 3D \((x_t, y_t, z_t)\) coordinates, where t is the specific frame. Given T number of time sequences and joint coordinates \(\textbf{v} = \{v_t \mid t=1, \cdots ,T\}\), we define nodes, edges, and angles as following:
where \(\mathcal {V}\) is a set of joints, \(\mathcal {E}\) and \(\mathcal {A}\) are sets of bones and angles defined by naturally connected two or three joints, respectively. To extract kinematic features, we calculated the \(L_2\)-norm of the n-th derivative of each element in these sets as follows:
where \(n=\{0, 1, 2\}\). Thus, we extract a total of 9 types of kinematic features summarized below:
2.3.2 Action Skeleton Details
We set our action skeletons to consist of 9 nodes, 8 edges, and 7 angles. To provide an intuitive understanding, we also provide specific nomenclature for defined nodes, edges, and angles, as below:
2.3.3 Evaluating Kinematic Similarity
To demonstrate the robustness of our proposed behavior embedding method, we quantitatively analyzed its ability to preserve kinematic relationships. By measuring the cosine similarity (\(\cos {\alpha }\)) between kinematic feature vectors and the normalized inter-point distance (\(\Delta x\)) in the embedding space, we could discern how closely the spatial arrangements of points (representing different behaviors) reflected their kinematic similarities (Fig. 3A).
Our analyses revealed that the UMAP-S particularly excels in reflecting kinematic relationships within its spatial configurations, as evidenced by a marked decline in kinematic feature similarity with increasing \(\Delta x\), which was more pronounced than with the t-SNE-S (Fig. 3B). Statistically significant contrasts between the UMAP and t-SNE techniques were further illuminated in Fig. 3C, showing UMAP-S enhanced capacity to correlate closer embedding distances with higher kinematic similarities. The first principal component (PC1) of kinematic features was then projected onto both t-SNE and UMAP embeddings, with UMAP-S demonstrating a more coherent distribution of kinematic information, thus visually confirming the statistical findings (Fig. 3D). In summary, our SUBTLE method does not merely capture temporal patterns but also adeptly structures kinematic features within the embedding space, with UMAP-S showing particular efficacy in aligning spatial proximity in the embedding with actual kinematic similarity, thereby offering a nuanced view of behavioral states.
2.4 Constructing Subclusters as Behavioral States
As a next step, we identified unique stereotyped actions (e.g., lifting, stretching, and balancing) as behavioral states (subclusters) (Berman et al., 2014) which can be used as building blocks for behavioral repertoires (superclusters) that reflect sequential transitions of behavioral states (e.g., rearing) (Berman et al., 2016). Several options are available to support this stage, including the Watershed algorithm (Berman et al., 2014), k-Means clustering (Menaker et al., 2022), Gaussian Mixture (Todd et al., 2017) and DBSCAN (Bodenstein et al., 2016). However, these algorithms require prior knowledge or assumptions about the embedding space, such as a probability density function, a predetermined number of clusters, or a threshold density. To overcome this, we used Phenograph clustering (Levine, 2015), a graph clustering algorithm that can cluster data points directly using Jaccard similarity and automatically determine the optimal number of clusters by maximizing the modularity of the clusters. Phenograph clustering produced better inner cluster inter-embedding kinematic similarities than k-Means clustering (Fig. 9), probably due to its ability to distinguish isolated groups in the embedding space (Fig. 9A). This demonstrates using the Phenograph clustering algorithm in SUBTLE leads to finding higher-quality clusters.
2.5 Drawing Biologically Meaningful Insights from Subclusters
We then utilized these subclusters to build a transition matrix between behavioral states. The state transition probabilities (shown by arrows in Fig. 4A) and the state retention rate (depicted by a hue in Fig. 4B) were used to determine the biological significance of these values. When comparing the retention rate with human-annotated labels (Fig. 4C), grooming behavioral states were commonly found in clusters with high retention rates, whereas walking or rearing was frequently found in clusters with relatively low retention rates (depicted by color in Figs. 4G and 11). While spectrogram-t-SNE gives similar patterns, the effect is more pronounced in the spectrogram-UMAP approach used in SUBTLE.
Next, we created a dendrogram using the similarity distance between animal subjects, calculated using the Frobenius norm of the behavior transition matrix (represented by color in Fig 4D, E). The transition matrix of behavioral states can be regarded as a distinct characteristic unique to each individual. By quantifying the similarity between these matrices, we can obtain valuable numerical data for comparing behavioral patterns across two distinct subjects. This approach allows for a more objective and measurable analysis of behavioral differences or similarities between individuals. The UMAP-based method distinguished young and adult mice better than the t-SNE method (Fig. 4F). Interestingly, data from the same subjects recorded in their young and old life stages (Fig. 10) appear close to each other when age information was removed (Fig. 4E). This result highlights that behavioral signatures based solely on the transition information between subclusters can capture the difference between young and adult subjects, yet the cases not distinctly separated belong to the same subject is biologically noteworthy. This result suggests that the information about state transitions of the subclusters created from the spectrogram-UMAP-based embedding space can be used to extract biologically meaningful insights without the need for human-provided behavior labels. In conclusion, we have developed Phenograph clustering and spectrogram-UMAP-based embeddings that can identify and interpret behavioral patterns in embedding space, which help extract biologically meaningful insights without human annotation.
2.6 Constructing Superclusters as Behavioral Repertoires
The prevailing theory of behavior suggests that it is organized in a temporally hierarchical manner. In line with this theory, we have constructed higher-level behavioral repertoires, which we refer to as superclusters (Berman et al., 2016). To obtain unsupervised behavior categories, we employed the predictive information-based deterministic information bottleneck (DIB) algorithm (Berman et al., 2016; Hernández, 2021; Strouse & Schwab, 2017). The algorithm aims to identify common patterns of an animal’s behavioral repertoires (superclusters) for accurate predictions of future behavioral states (subclusters), while minimizing reliance on past behavioral states by finding the optimal trade-off between predictability of the future and reliance on past. We grouped the subclusters into n superclusters that contained information about the future state after \(\tau \) steps while retaining less information about the past state (See Superclustering method section in Method 5.4.4 for more details). As a result, we identified superclusters for n = 2, 4, and 6 (Fig. 5A) and human-annotated labels have similar structures to the n = 6 case (See Fig. 5B). For n = 6, the transition matrix of subclusters with grouping by the obtained superclusters is shown in Fig. 5C. Clustering results and the grouped transition matrix for cases where n ranges from 1 to 7 can be found in fig. S5B and S6C. A human-interpretable hierarchical structure can be observed in the Sankey diagram flow (See Fig. 5D) as n increases. An example of the 30s animal behavior, an ethogram, is also presented for n = 2, n = 6, subcluster, human-annotated cases in Fig. 5E.
We assessed the accuracy of the superclusters from the DIB algorithm compared to human-annotated animal behavior labels (Fig 5F) using Normalized Mutual Information (NMI) (Strehl & Ghosh, 2002) and the Adjusted Rand Index (ARI) (Hubert & Arabie, 1985; Steinley, 2004), two widely used metrics for evaluating the performance of unsupervised clustering algorithms. We show that TSNE-S-Pheno-DIB, achieves similar performance compared to MotionMapper-DIB, while SUBTLE exhibits twice higher performance gains than others, as the number of superclusters (n) increases. (See Figs. 5F and 13). We also confirmed the actual clustering result by viewing a recorded video, 3D action skeleton, human annotation, and SUBTLE supercluster simultaneously Fig. 14.
2.7 Drawing biologically meaningful insights from superclusters
After confirming the accuracy of superclusters, we further perform a detailed analysis to find out new biological meanings. To extract biologically meaningful insights from superclusters, we evaluated age-dependent behavioral changes by analyzing the differentially exhibited kinematic features (young vs. adult mice; Fig. 5G). We found most of the significant differences from zero derivatives of edges in all 6 superclusters, which may be due to the varying size between young and adult mice (Fig. 5G). Except for zero derivatives of edges, the majority of differences were found in rearing (17 features) and walking (9 features) clusters (Fig. 5G), indicating that young and adult mice have substantial differences in terms of dynamic movement. The smallest difference between young and adult mice was found in grooming-like behaviors (superclusters 1 and 2). The proportion of significantly different features is distinct for each supercluster (Fig. 5H). While our differential analysis of inter-group kinematic features provides interpretable enriched data, the conventional approach that estimates locomotor activity by 2D-trajectory distances in an open-field-like environment revealed no significant inter-group difference (fig. S3C and D). In conclusion, by combining superclusters and kinematic feature analysis, our method successfully distinguishes subtle behavioral changes between young and adult mice.
2.8 Temporal information is critical for behavioral categorization
We investigated the effect of random shuffles on the sequence of behavioral states to better understand the role of temporal connectivity in behavioral categorization. Specifically, we divided the sequence of behavioral states into equal-sized chunks and randomly swapped the order of the chunks to create a shuffled sequence (See Fig. 6A for an illustration with chuck size = 4). We then measured the effect of shuffling on the temporal connectivity of the data using the TPI. We found that as the chunk size decreased, the TPI metric was disrupted, indicating no temporal connectivity in the shuffled data (See Fig. 6B). Additionally, we evaluated the effect of shuffling on the performance of the superclustering algorithm using ARI and NMI scores. We found that as the chunk size decreased, ARI and NMI scores were also disrupted (See Fig. 6C). These results indicate that our TPI metric effectively captures temporal dynamics information and underscores the importance of temporal connectivity in behavioral categorization.
2.9 SUBTLE as a universal behavior mapping framework
To demonstrate the general usability of the SUBTLE framework, we applied our method in various publicly available datasets from mouse (Shank3KO) (Huang, 2021), rat (DANNCE) (Dunn, 2021), and human (Fit3D) (Fieraru et al., 2021). We first investigate the temporal connectivity patterns in these datasets. We show that the UMAP-S embedding excels in the TPI scores compared to the t-SNE-S, regardless of the species or recording methods (Fig. 7B, D, F). Since the Fit3D dataset provides 47 categorical labels, we visualized them in the embedding space (Fig. 7A). As can be seen in the figure, UMAP-S seems better at preserving global structure compared to t-SNE-S, represented in a distance in the embedding space (e.g. walk-the-box). By measuring the clustering performance of subclusters, we show that UMAP-S in combination with Phenograph clustering shows the best performance compared to other methods (Fig. 7C), highlighting the effectiveness of using both UMAP-S and Phenograph algorithms in the SUBTLE framework. Due to lack of the human-annotated labels in Shank3KO and DANNCE datasets, with the identification of both subclusters and superclusters with SUBTLE, we have visually inspected the clustering quality by using a web-based user-friendly GUI tool (Fig. 14 and sVideo. 1). Using Shank3KO dataset, we found the impressive mapping of animal behaviors such as jumping behavior in subcluster 78 (sVideo. 2). Together, we emphasize the universal applicability of the SUBTLE framework in various species and acquisition environments.
3 Conclusion
This paper introduced the TPI, a novel approach that combines sequential data with k-Means clustering to measure the temporal structure by evaluating the transition probability between arbitrary k clusters and the distances between cluster centers. A critical aspect of this approach is the validity of comparisons, which is only ensured when the embedding space has the same k value across the datasets being compared.
In terms of feature embedding methods, we found that using spectrograms preserves temporal structures more effectively than other feature extraction techniques, such as raw coordinates or kinematic features. Furthermore, when performing nonlinear embedding, UMAP outperformed t-SNE in capturing temporal structures more accurately. As a key finding, our data suggests that a combination of spectrograms and UMAP not only preserves the temporal structure but also effectively maintains the kinematic structure in animal behavioral analysis. This unique combination produced clustering results akin to human-annotated labels, demonstrating its effectiveness and accuracy.
In sum, we conclude that the proposed SUBTLE framework can effectively utilize the information from subclusters and superclusters to distinguish biologically significant characteristics (e.g., Young and Adult groups in mice). We expect that SUBTLE, with its reduced human bias, user-friendly interface, and robust validation metric (TPI), has the potential to become a useful public resource for the broader research community in computer vision and animal behavior analysis.
4 Discussion
Despite the success of deep learning in detecting 3D points in behaving animals, accurately identifying higher-order behavioral patterns remains a challenge for machine learning systems. While humans can contribute and classify animal behaviors using their intuition and labeling guidelines, incorporating human annotation into artificial behavioral analysis is labor-intensive and prone to observer bias. This limitation exists especially for large datasets, and the results may be influenced by the subjective judgments of the annotators. As a result, researchers have sought alternative approaches to overcome these roadblocks, including unsupervised methods that do not require human-provided labels. However, this approach raises a dilemma: the outcomes may differ from those of humans as the model does not have constraining or intuitive guidelines.
In this study, we developed a novel method called SUBTLE (spectrogram-UMAP-based temporal-link embedding) to map and analyze animal behavior. We evaluated the quality of behavioral embedding space by measuring the temporal representation using TPI. Our results show that using the wavelet spectrogram in combination with UMAP results gave the best temporal connectivity, outperforming other feature embedding methods. Additionally, SUBTLE effectively reflects kinematic similarity as a distance more effectively than other approaches. Our findings suggest that SUBTLE can help analyze animal behavior, as demonstrated in the analysis of \(\sim \)190-minute-long logs (\(\sim \)228,000 frames, 19 mice) of mice movements and the corresponding \(\sim \)100-minute-long logs (\(\sim \)120,000 frames, 10 mice) of human annotations. Furthermore, our study highlights the importance of temporal dynamics information in behavioral analyses and the value of using feature embeddings to extract meaningful information from raw data.
One of the key contributions of our study is the suggestion of an evaluation strategy, including the development of the TPI metric to assess temporal connectivity in the feature embedding space. This allows for a more rigorous evaluation of the method and provides insight into how to construct a behavioral embedding space that enables machines to categorize animal behavior similar to humans. Moreover, the proposed hypothesis on the human brain using transition probabilities for sequence categorization well matches the concept of behavioral state transitions in the SUBTLE framework. Our proposed TPI metric and kinematic similarity measurement can be used for explicit constraints for setting learning objectives in AI-based modeling. The recent successes of large language models in natural language processing suggest the exciting possibility of creating large behavior models that could advance our understanding of animal behavior. These possibilities await future investigation by researchers, and we look forward to the potential breakthroughs that may arise from these developments.
One of the strengths of the TPI is its ability to measure the temporal structure within low-dimensional embeddings. This measurement can represent the degree of temporal distortion in the embedding space, with higher TPI indicating less distortion and lower TPI indicating more distortion. This aspect of TPI is particularly relevant in the ongoing debates (Vogelstein, 2021; Sousa & Small, 2022) about how to effectively and accurately embed high-dimensional spatiotemporal data into lower dimensions. Our research contributes to this debate by seeking to balance between practical utility and analytical accuracy in dimensionality reduction, with a particular emphasis on temporal representation. Recent discussions stress the importance of cautious use of dimensionality reduction techniques, as improper application can lead to distorted outcomes like phantom oscillations in the time-space representation (Shinn, 2023; Dyer & Kording, 2023). To conclude, our findings contribute to the ongoing debate on dimensionality reduction by highlighting the TPI as a promising tool for ensuring minimal temporal distortion in lower-dimensional representations of complex spatiotemporal data.
Our SUBTLE framework offers several advantages for researchers. Firstly, it saves a significant amount of time by automating the identification of behavior repertoires, eliminating the need for labor-intensive manual annotation. Secondly, it is free from human subjective bias in the behavioral labeling process, resulting in a more reliable and unbiased identification of behavior repertoires. Thirdly, the framework allows for the identification of individual behavior signatures, which can be used for comparison with other individuals or groups (Fig. 4D-F). Finally, it can profile inter-group subtle kinematic feature differences, providing valuable insights into the nuances of animal behavior (Fig. 5G and H). These advantages make SUBTLE a valuable tool for advancing our understanding of animal behavior and potentially saving time and resources for researchers. Our spectrogram-based unsupervised framework and mouse action skeleton data will be released to the public (See Data and Materials availability in Acknowledgement).
5 Materials and Methods
5.1 Animals
Young (3–4 weeks) and adult (8–12 weeks) Balb/c male mice (IBS Research Solution Center) were used for the acquisition of action skeletons. Every animal experimental procedure was approved by the Institutional Animal Care and Use Committee of the Institute for Basic Science (IBS, Daejeon, Korea). The animals were kept on a 12-hour light–dark cycle with controlled temperature and humidity and had ad libitum access to food and water.
5.2 Behavior recording procedure
Recordings of behavior were performed after 3 days of handling and habituation. In order to reduce background stress and anxiety in the laboratory mice, cotton gloves were used during the whole handling process. At least 2 h before the experiment, animals were moved to the experimental room for habituation. The spontaneous animal movements were recorded using the AVATAR system (Kim et al., 2022) for 10 min for 10 min/day on 5 consecutive days. In this study, we utilized the first day of recordings from all mice. Between each session, the inner transparent recording chamber was cleaned with 70 % ethanol and dried thoroughly. To prevent the effect of noise, the whole recording system was in a sound-attenuated room.
5.3 Data Acquisition
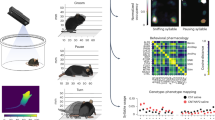
We collected action skeletons with AVATAR, a YOLO-based (Redmon et al., 2015; Redmon & Farhadi, 2018) 3D pose estimation system with multi-view images that extracts continuous joint movement from freely moving mice (Kim et al., 2022). The AVATAR system, with its utilization of five cameras, exhibits robustness in handling occlusion. Particularly, the inclusion of a bottom-view camera enables accurate tracking of limbs, resulting in increased freedom from occlusion compared to the top view. As previously reported (Kim et al., 2022), the network shows an Intersection over Union (IOU) score exceeding 75%. For Mean Average Precision (mAP), AVATARnet accurately detected nine body parts at a rate of 90%. Post-training, each class presented a Mean Square Error (MSE) distance between 7 and 15 pixels (1.4–4.5 mm). Moreover, the center points of detected body objects were found to align closely with locations marked by human observers. This system is commercially available at https://www.avataranalysis.com/.
The recording hardware consists of an inner transparent chamber and an outer recording box. The outer recording box consists of 5 Full-HD CMOS cameras (FLIR, U3-23S3M/C-C, 1200\(\times \)1200 pixels, 20 frames per second): four at the side and one at the bottom, with a distance of 20 cm (side), and 16 cm (bottom) from the inner transparent chamber. These cameras were directly connected to the recording computer (CPU, Intel, i9-10900K; GPU, Colorful, RTX3080; RAM, 16GB). As high-resolution image acquisition is required to detect an animal’s joint coordinates accurately, the recording was conducted under bright lighting conditions. The brightness of the lighting inside the recording box was 2950 lux. The inner transparent chamber is positioned at the center of the outer recording box. For the inner transparent recording chamber, the size of the cuboid is 30 cm \(\times \) 20 cm \(\times \) 20 cm (height \(\times \) width \(\times \) length). Numerous holes with a diameter of 5 mm were made on the upper side of the chamber in order not to impede the animal’s breathing. The top of the transparent inner chamber is covered with a white background lid after the animal enters. Acquired video files are processed with a pre-trained YOLO-based DARKNET model to extract the coordinates of mouse joints.
For camera calibration, we follow the instructions from the manufacturer (ACTNOVA) (Kim et al., 2022). Briefly, based on “triangulate function” and “vision toolbox” in MATLAB, we have established an automated method for calibrating cameras using videos captured concurrently of a standard calibration board (checkerboard pattern) that is manually moved across the view of the cameras (Fig. 15). Using the parameters of each camera (intrinsic, extrinsic, and lens distortion), we successfully achieve the 3D coordinate of body parts from multiple 2D images.
5.4 Mapping Behavioral States
Our method is inspired by previous spectrogram-based behavior mapping approach (Berman et al., 2014, 2016). Here, we describe the details of the method.
5.4.1 Continuous Wavelet Transformation
First, we extracted C channels of spectrogram via wavelet transformation for given D euclidean dimensions and N nodes; D and N were 3 and 9, respectively. We used node information only. We employed the Morlet wavelet (Goupillaud et al., 1984), which has the following formulation:
and the continuous wavelet transformation (CWT) of signal x is calculated via
where \(\omega _0\) is the central frequency of the wavelet, s is the scale parameter, and t is time. \(\omega _0\) determines the dominant frequency content of the Morlet wavelet, while s determines the effective time window of the wavelet. The time scale s and the fundamental frequency f have the following relationship:
where \(f_s\) is the sampling rate. After the wavelet transformation, the power spectrum S(f; t) (i.e., wavelet spectrogram) is calculated as
We set \(\omega _0=5\) and the frequency range was given as [\(f_\text {min}\), \(f_\text {max}\)], \(f_\text {max}=f_s/2\) Hz (Nyquist frequency), and \(f_\text {min}=f_\text {max}/10\) Hz. In our experiment, \(f_s\) was 20 Hz so that \(f_\text {max}\) and \(f_\text {min}\) were set at 10 Hz and 1 Hz, respectively. The frequency interval between \(f_\text {max}\) and \(f_\text {min}\) was evenly spaced into 50 segments, i.e. \(C=50\).
Spectrograms were obtained for each node and Euclidean dimension axis. Thus, the preprocessed features were given as multiple spectrograms, and the dimension of the feature at time t was \(D \times N \times C\). Including the original raw Euclidean coordinates \(D \times N\), the final feature dimension is \(D \times C \times (N+1) = 3 \times 50 \times 10 = 1500\). Here, D represents the Euclidean dimension, C represents the number of spectrogram channels, and N represents the number of keypoints or nodes. The total number of frames per session was 12,000, and the number of sessions was 10. We applied the CWT for each session; after the CWT, we randomly sampled 12000 data points and applied the principal component analysis (PCA) on this randomly selected dataset due to its computational efficiency. After PCA, we selected the top 100 principal components (PCs) and mapped all preprocessing data (i.e., CWT data) to these 100 PCs. By first reducing the dimensionality of the data using PCA, the computation time for UMAP or t-SNE can be reduced, especially when dealing with high-dimensional datasets.
5.4.2 Nonlinear Embedding Methods
Next, we conducted nonlinear dimension reduction into 2-dim from 100 PCs for clustering purposes. This 2-dim embedding space is called behavior space. We demonstrated two nonlinear embedding methods t-SNE (Van der Maaten & Hinton, 2008) and UMAP (McInnes et al., 2018), for creating behavior spaces. \(z_t\) denotes a point at time step t in the behavior space. The number of neighbors in the UMAP method was set to 50. The perplexity of t-SNE was set at 30.
5.4.3 Subclustering Methods
Applying UMAP or t-SNE to the reduced data can further enhance the clustering results by revealing more structure in the data. With the given set of points in the behavior space or embedding space, we performed PhenoGraph clustering (Levine, 2015) and k-Means clustering methods to determine k communities or subclusters.
PhenoGraph clustering was originally designed for high-dimensional single-cell phenotyping. In this method, each node or point in the behavior space is connected to its \(\kappa \) nearest neighbors in the behavior space. Thereby, a \(\kappa \)-nearest neighbor graph is created. After that, PhenoGraph uses modularity optimization (e.g., Louvain or Leiden) to partition the graph into k communities or subclusters of nodes that are more similar to each other than nodes in other subclusters. We utilized the Leiden method, which produces results more quickly than Louvain (Traag et al., 2019). The modularity of the partitioned graph is maximized when nodes are more densely connected within the same subcluster than to other nodes in different subclusters. If transitions between nodes are more likely to happen when the two nodes are closely located in the embedding space than when they are separated far away, the trajectory in the behavior space would indicate staying longer in the same community. We call this community or subcluster behavior state S(t); the transition matrix \(\textbf{T}(n)\) is defined on this behavior state, where n is the number of steps between two states. We set \(\kappa \) to 30 in the PhenoGraph algorithm. The PhenoGraph algorithm requires no predefined number of communities k to automatically find the optimal k for the modularity of the subdivided graph.
In contrast, the objective of k-Means clustering is to divide the entire dataset into k clusters such that the sum of squared distances between each point and the cluster center is minimized. In this method, modularity is not taken into account, and k must be predetermined.
5.4.4 Superclustering Method
After the subclustering process, we employed the predictive information bottleneck method for obtaining superclusters. Berman et al. (2016) showed that (1) the transition matrix of flies’ behavior states has hierarchical structures and (2) a sequence of behavioral states has a longer time scale than the Markovian assumption, i.e., history of behavior states affects the next behavior transition. Based on this observation, they proposed dividing behavioral states into superclusters that maintain a significant amount of the information regarding future actions (predictive information) that is provided by the current behavioral state.
In a Markovian system, the mutual information between two behavior states in time \( t_1 \) and \( t_2 \), \( I(S(t_1), S(t_2)) \), decays exponentially for large separation \( |t_2-t_1 |\) and a characteristic decay time is
where \( \lambda _2 \) is the second largest eigenvalue of \(\textbf{T}(n=1) \). We grouped our behavior states S(t) into groups Z such that maintains the information about the future state after \(2t_c\) transition steps (Berman et al., 2016). In detail, we maximized the following equation:
where \(\tau \) is \(2t_c\), the first term increases the information about the state after \(\tau \) transitions in the future, while the second term reduces information from the past; \(\beta \) is a parameter that controls the predictive power of the supercluster Z. The deterministic information bottleneck (DIB) algorithm was utilized to optimize this equation, and the optimization procedure followed was the same as in (Berman et al., 2016).
Our model is a sequential combination of CWT, PCA, UMAP (or t-SNE), PhenoGraph (or k-Means), and predictive information bottleneck. For the new dataset, we used a pretrained model for behavioral state mapping. Note that for new embedding \(\hat{z}\), we used the nearest neighbor on the training dataset with previously detected subclusters or communities in the PhenoGraph stage.
5.5 Temporal Connectivity Analysis of Behavioral State Embeddings
To evaluate the temporal connectivity of nonlinear state embeddings, we generated discrete behavioral state categories by conducting k-Means clustering while increasing k. Next, we evaluated the TPI using an inter-cluster transition probability matrix and cluster centroids.
5.6 Comparison of Superclusters and Human-Annotated Categories
We used two evaluation metrics: Adjusted Rand Index (ARI) (Hubert & Arabie, 1985; Steinley, 2004) and Normalized Mutual Information (NMI) (Strehl & Ghosh, 2002). The NMI ranges between 0 and 1, and the ARI ranges between -1 and 1. For all metrics, the higher the better, and all of them can accommodate different numbers of classes between the superclusters and the human-annotated categories.
5.7 Manual Annotation for Behavior Categories
Annotation of behavior was performed manually by trained researchers using a custom-built GUI interface. Researchers were instructed to annotate each video frame with one of four predefined categories of behavior: walking, grooming, rearing, and standing. “Walking” indicates when the mouse shows movement, specifically with changes in the paw positions. “Grooming" indicates when the mouse strokes the forepaw(s) or licks the body parts. “Rearing" is when the mouse puts weight on its hind legs and lifts its forepaws off the ground. “Standing" is when all of the mouse’s paws are on the ground and it is standing still. “na" is the abbreviation for not annotated.
5.8 Mapping Animal Behavior with MotionMapper
MotionMapper libraryFootnote 1 offers various nonlinear mapping techniques, including not only t-SNE but also Variational Auto Encoder (VAE) and UMAP for behavioral state embeddings. The original work only focused on the PCA component of the video, whereas the keypoint trajectory data can also be handled using VAE. Therefore, we have adopted the following embedding protocol suggested by the motion mapper library, which encompasses these additional techniques:
Keypoint trajectory data (9 key points \(\times \) 3D = 27 dim in total) \(\rightarrow \) VAE (projecting to latent space; 8 dim) \(\rightarrow \) Wavelet transform (8 \(\times \) 30 (wavelet frequencies from 1Hz to 10Hz) = 240 dim in total) \(\rightarrow \) UMAP (projecting to 2 dim) \(\rightarrow \) Watershed (subcluster; 20 behavioral states) \(\rightarrow \) Deterministic Information Bottleneck (supercluster; number of supercluster = 1–7)
5.9 Computational Cost of SUBTLE Framework
To aid future studies for running the SUBTLE framework, we provide computational cost in terms of time and memory usage. As can be seen in the Fig. 17, the major time cost in the SUBTLE framework is Phenograph clustering, and the memory cost is the CWT for spectrogram generation. However, since this evaluation result assumes the frames come from single-take recording, it will differ based on the actual dataset configuration such as the total number of animals and the length of frames in each recording. While our framework is designed for only CPU configuration, future works using GPUs have the potential to improve the training speed, while this possibility is not addressed in this work yet.
5.10 Training Details for Public Datasets
For evaluation of the Fit3D dataset (Fieraru et al., 2021), we used 3D skeletons provided in training datasets,Footnote 2 after obtaining permission to use the dataset. For training with the SUBTLE framework, the framerate was set to 50Hz, and both raw coordinates and spectrograms were used for input features. For visualization, we showed the valid annotation by filtering with ‘Repetition Segmentations’, which is provided in the dataset.
For the DANNCE dataset, we used the first 30-minute frames for each recording from Rat_7M dataset Footnote 3 (total 378,000 = 54,000\(\times \)7 frames), following the original paper. For training with the SUBTLE framework, the framerate was set to 30Hz, and both raw coordinates and spectrograms were used for input features. As videos provided by authors are 4\(\times \) speed and segmented, we have converted to normal speed and concatenated the videos for GUI visualization.
For the Shank3KO dataset ,Footnote 4 it is composed of 20 recordings with 15-minute lengths with 30Hz framerate (total 540,000 = 27,000\(\times \)20 frames). For training, both raw coordinates and spectrograms were used for input features. The distribution of preprocessed data used in our study can be available upon request with the original author’s permission.
3D action skeleton acquisition system. A Action skeleton extraction using 3D pose estimation system. Illustration of recording environment and data extraction steps of AVATAR system (left). B captured snapshots of the subject mouse from 5-way cameras. C reconstructed representative action skeletons of various actions with joint numbers and names. D Example reconstructed 3D skeletons are depicted for 4 behavior categories
Phenograph clustering outperforms k-Means clustering for subcluster generation for SUBTLE. A Subclusters detected by k-Means clustering. Yellow arrows indicate the clusters that contain more than two visually isolated chunks. B Subclusters detected by Phenograph clustering. C Comparison of inner-cluster mean kinematic similarity between k-Means clustering and PhenoGraph clustering with 20 trials. D Summary bargraph of inner-cluster mean kinematic similarity. (p<0.001, Welch’s t-test) (Color figure online)
State-transition and retention map of subclusters constructed by k-Means clustering. A State retention map of Phenograph subclusters in embeddings of spectrogram t-SNE and spectrogram UMAP. B State transition map of Phenograph subclusters in embeddings of spectrogram t-SNE and spectrogram UMAP. C Empirical cumulative distribution function of human-annotated behavioral states against retention rate
Transition probabilities and behavioral modularity for SUBTLE-Pheno-DIB. A Markov model transition matrix for \(\tau =40\) (left), Transition matrices for \(\tau =40\) (center) and \(\tau =400\) (right). B Information bottleneck clustering of SUBTLE space for \(\tau =20\) (approximately twice the longest time scale in the Markov model). C One-step Markov transition probability matrix \(\tau =1\). The 89 subclusters are grouped into n number of superclusters by applying the predictive information bottleneck calculation. White lines denote the cluster boundaries
Comparison of clustering quality between SUBTLE-Pheno-DIB and SUBTLE-K-Means method. A Behavior categories depicted on embedding space by SUBTLE-K-Means method (top) SUBTLE-Pheno-DIB (bottom). B Sankey Diagram of SUTBLE-Pheno-DIB. C Adjusted rand index scores calculated by comparing with human-annotated categories. D Normalized mutual information calculated by comparing with human-annotated categories
SUBTLE visualization framework in the website (https://subtle.ibs.re.kr). A 3D action skeleton visualization by Three.js. B Visualization of current behavior status (human annotation or SUBTLE supercluster-cluster). C Actual video of mouse behavior taken from AVATAR. D Controller panel. Fps: Play speed of video; Trace: The number of frames remaining in 3D action skeleton; Frameskip: The number of frames for skipping; AutoRotation: Apply for slow rotation in 3D skeleton; Playstatus: For play or pause the video; Darkskin: Change the color mode (black background) in 3D skeleton; Time: The number of the current frame. E Annotation filter. (top) “Human annotation” or “SUBTLE supercluster.” (bottom) Checkbox for each annotation label or the number of superclusters. F The result of behavior mapping after SUBTLE
Computational cost of SUBTLE algorithm. A Time cost in minutes (min) with a varying number of trained frames. Colored areas represent the proportional time cost within the total time cost in the SUBTLE algorithm (gray scatter plot). B Memory cost in gigabytes (GB) with a varying number of trained frames. Scatter plots represent the usage of memory while running the various algorithms, while the gray area represents the maximum usage of memory in the SUBTLE algorithm (equivalent to CWT memory usage). In summary, the Phonograph (red) is the dominant bottleneck in terms of time cost and CWT (blue) is the dominant bottleneck in terms of the memory cost in SUBTLE (Color figure online)
5.11 Developing visualization homepage (subtle.ibs.re.kr)
We found that a web application, such as a homepage, could be an ideal platform for simultaneous visualization of coordinates, videos, and behavioral labels due to its computing speed and versatility. To visualize the 3D coordinates of animals, we used the "Three.js" library. To visualize embedding results from SUBTLE, we adopted the "d3js", which is a fast plotting library. Then we synchronize coordinates, videos, and labels based on the frame number of videos. The visualization of other datasets, including Shank3KO and DANNCE, can also be achieved by adapting the configuration settings for coordinates (Fig. 16).
References
Bala, P. C., et al. (2020). Automated markerless pose estimation in freely moving macaques with openmonkeystudio. Nature Communications, 11(1), 4560.
Berman, G. J., Bialek, W., & Shaevitz, J. W. (2016). Predictability and hierarchy in drosophila behavior. Proceedings of the National Academy of Sciences, 113(42), 11943–11948.
Berman, G. J., Choi, D. M., Bialek, W., & Shaevitz, J. W. (2014). Mapping the stereotyped behaviour of freely moving fruit flies. Journal of The Royal Society Interface, 11(99), 20140672.
Bodenstein, C., Götz, M., Jansen, A., Scholz, H., & Riedel, M. (2016). Automatic object detection using dbscan for counting intoxicated flies in the Florida assay (pp. 746–751).
Bohnslav, J. P., et al. (2021). Deepethogram, a machine learning pipeline for supervised behavior classification from raw pixels. Elife, 10, e63377.
Brattoli, B., et al. (2021). Unsupervised behaviour analysis and magnification (uBAM) using deep learning. Nature Machine Intelligence, 3(6), 495–506.
Cande, J., et al. (2018). Optogenetic dissection of descending behavioral control in drosophila. Elife, 7, e34275.
Chang, E. F., et al. (2010). Categorical speech representation in human superior temporal gyrus. Nature Neuroscience, 13(11), 1428–1432.
De Oliveira, M. F., & Levkowitz, H. (2003). From visual data exploration to visual data mining: A survey. IEEE Transactions on Visualization and Computer Graphics, 9(3), 378–394.
DeAngelis, B. D., Zavatone-Veth, J. A., & Clark, D. A. (2019). The manifold structure of limb coordination in walking drosophila. Elife, 8, e46409.
Dehaene, S., Meyniel, F., Wacongne, C., Wang, L., & Pallier, C. (2015). The neural representation of sequences: From transition probabilities to algebraic patterns and linguistic trees. Neuron, 88(1), 2–19.
Dunn, T. W., et al. (2021). Geometric deep learning enables 3d kinematic profiling across species and environments. Nature Methods, 18(5), 564–573.
Dyer, E. L., & Kording, K. (2023). Why the simplest explanation isn’t always the best. Proceedings of the National Academy of Sciences, 120(52), e2319169120.
Fieraru, M., Zanfir, M., Pirlea, S. C., Olaru, V., & Sminchisescu, C. (2021). Aifit: Automatic 3d human-interpretable feedback models for fitness training, pp. 9919–9928.
Goupillaud, P., Grossmann, A., & Morlet, J. (1984). Cycle-octave and related transforms in seismic signal analysis. Geoexploration,23(1), 85–102.
Graving, J. M., & Couzin, I. D. (2020). VAE-SNE: a deep generative model for simultaneous dimensionality reduction and clustering. BioRxiv 2020–07.
Günel, S., et al. (2019). Deepfly3d, a deep learning-based approach for 3d limb and appendage tracking in tethered, adult drosophila. Elife, 8, e48571.
Hernández, D. G., et al. (2021). A framework for studying behavioral evolution by reconstructing ancestral repertoires. Elife, 10, e61806.
Hsu, A. I., & Yttri, E. A. (2021). B-SOiD, an open-source unsupervised algorithm for identification and fast prediction of behaviors. Nature Communications, 12(1), 5188.
Huang, K., et al. (2021). A hierarchical 3d-motion learning framework for animal spontaneous behavior mapping. Nature Communications, 12(1), 1–14.
Hubert, L., & Arabie, P. (1985). Comparing partitions. Journal of classification, 2, 193–218.
Johansson, G. (1973). Visual perception of biological motion and a model for its analysis. Perception & Psychophysics, 14(2), 201–211.
Karashchuk, P., et al. (2021). Anipose: A toolkit for robust markerless 3d pose estimation. Cell Reports, 36(13), 109730.
Kim, D.-G., Shin, A., Jeong, Y.-C., Park, S., & Kim, D. (2022). Avatar: AI vision analysis for three-dimensional action in real-time. bioRxiv 2021–12.
Klaus, A., et al. (2017). The spatiotemporal organization of the striatum encodes action space. Neuron, 95(5), 1171–1180.
Kobak, D., & Linderman, G. C. (2021). Initialization is critical for preserving global data structure in both t-SNE and UMAP. Nature Biotechnology, 39(2), 156–157.
Kojima, T., Gu, S. S., Reid, M., Matsuo, Y., & Iwasawa, Y. (2022). Large language models are zero-shot reasoners. Advances in Neural Information Processing Systems, 35, 22199–22213.
Lake, B. M., Salakhutdinov, R., & Tenenbaum, J. B. (2015). Human-level concept learning through probabilistic program induction. Science, 350(6266), 1332–1338.
Levine, J. H., et al. (2015). Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell, 162(1), 184–197.
Luxem, K., et al. (2022). Identifying behavioral structure from deep variational embeddings of animal motion. Communications Biology, 5(1), 1267.
Marks, M., et al. (2022). Deep-learning-based identification, tracking, pose estimation and behaviour classification of interacting primates and mice in complex environments. Nature Machine Intelligence, 4(4), 331–340.
Marques, J. C., Lackner, S., Félix, R., & Orger, M. B. (2018). Structure of the zebrafish locomotor repertoire revealed with unsupervised behavioral clustering. Current Biology, 28(2), 181–195.
Marshall, J. D., et al. (2021). Continuous whole-body 3d kinematic recordings across the rodent behavioral repertoire. Neuron, 109(3), 420–437.
McInnes, L., Healy, J., & Melville, J. (2018). Umap: Uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426.
Mearns, D. S., Donovan, J. C., Fernandes, A. M., Semmelhack, J. L., & Baier, H. (2020). Deconstructing hunting behavior reveals a tightly coupled stimulus-response loop. Current Biology, 30(1), 54–69.
Menaker, T., Monteny, J., de Beeck, L. O., & Zamansky, A. (2022). Clustering for automated exploratory pattern discovery in animal behavioral data. Frontiers in Veterinary Science, 9, 884437.
Nath, T., et al. (2019). Using deeplabcut for 3d markerless pose estimation across species and behaviors. Nature Protocols, 14(7), 2152–2176.
Nilsson, S. R., et al. (2020). Simple behavioral analysis (simba)–an open source toolkit for computer classification of complex social behaviors in experimental animals. BioRxiv 2020–04.
Pereira, T. D., et al. (2019). Fast animal pose estimation using deep neural networks. Nature Methods, 16(1), 117–125.
Pereira, T. D., Shaevitz, J. W., & Murthy, M. (2020). Quantifying behavior to understand the brain. Nature Neuroscience, 23(12), 1537–1549.
Redmon, J., & Farhadi, A. (2018). Yolov3: An incremental improvement. arXiv preprint arXiv:1804.02767.
Redmon, J., Divvala, S., Girshick, R., & Farhadi, A. (2015). You only look once: unified, real-time object detection. arXiv preprint arXiv:1506.02640.
Segalin, C., et al. (2021). The mouse action recognition system (MARS) software pipeline for automated analysis of social behaviors in mice. Elife, 10, e63720.
Shima, K., Isoda, M., Mushiake, H., & Tanji, J. (2007). Categorization of behavioural sequences in the prefrontal cortex. Nature, 445(7125), 315–318.
Shinn, M. (2023). Phantom oscillations in principal component analysis. Proceedings of the National Academy of Sciences120(48), e2311420120. https://www.pnas.org/doi/abs/10.1073/pnas.2311420120. https://doi.org/10.1073/pnas.2311420120, https://www.pnas.org/doi/pdf/10.1073/pnas.2311420120.
Sousa, D., & Small, C. (2022). Joint characterization of spatiotemporal data manifolds. Frontiers In Remote Sensing, 3, 760650.
Steinley, D. (2004). Properties of the Hubert-Arable adjusted rand index. Psychological Methods, 9(3), 386.
Strehl, A., & Ghosh, J. (2002). Cluster ensembles–a knowledge reuse framework for combining multiple partitions. Journal of machine learning research, 3(Dec), 583–617.
Strouse, D., & Schwab, D. J. (2017). The deterministic information bottleneck. Neural Computation, 29(6), 1611–1630.
Sturman, O., et al. (2020). Deep learning-based behavioral analysis reaches human accuracy and is capable of outperforming commercial solutions. Neuropsychopharmacology, 45(11), 1942–1952.
Todd, J. G., Kain, J. S., & de Bivort, B. L. (2017). Systematic exploration of unsupervised methods for mapping behavior. Physical Biology, 14(1), 015002.
Traag, V. A., Waltman, L., & Van Eck, N. J. (2019). From Louvain to Leiden: Guaranteeing well-connected communities. Scientific Reports, 9(1), 1–12.
Van der Maaten, L., & Hinton, G. (2008). Visualizing data using t-SNE. Journal of Machine Learning Research, 9(11), 2579–2605.
Vogelstein, J. T., et al. (2021). Supervised dimensionality reduction for big data. Nature Communications, 12(1), 2872.
Weinreb, C., et al. (2023). Keypoint-moSeq: parsing behavior by linking point tracking to pose dynamics. BioRxiv 2023–03.
Willmore, L., Cameron, C., Yang, J., Witten, I. B., & Falkner, A. L. (2022). Behavioural and dopaminergic signatures of resilience. Nature, 611(7934), 124–132.
Wiltschko, A. B., et al. (2015). Mapping sub-second structure in mouse behavior. Neuron, 88(6), 1121–1135.
York, R. A., Carreira-Rosario, A., Giocomo, L. M., & Clandinin, T. R. (2020). Flexible analysis of animal behavior via time-resolved manifold embedding. BioRxiv 2020–09.
Zhang, Y., Han, K., Worth, R., & Liu, Z. (2020). Connecting concepts in the brain by mapping cortical representations of semantic relations. Nature Communications, 11(1), 1877.
Zhou, Y., et al. (2022). Large language models are human-level prompt engineers. arXiv preprint arXiv:2211.01910.
Zimmermann, C., Schneider, A., Alyahyay, M., Brox, T., & Diester, I. (2020). Freipose: A deep learning framework for precise animal motion capture in 3d spaces. BioRxiv 2020–02.
Acknowledgements
The authors would like to thank Dae-Gun Kim and Jungjoon Park from ACNTNOVA for their helpful discussions and experimental support on the AVATAR system.
Funding
Open Access funding enabled and organized by KAIST. This work was supported by the Institute for Basic Science (IBS), Republic of Korea. D. K. K. and M. C. were supported by IBS under Grant Number IBS-R029-C2. J. K., S. K., J. H. J., S. H. K., and C. J. L. were supported by IBS under grant number IBS-R001-D2. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: JK, SK; Data curation: JK, SK, JJ, SHK; Methodology: JK, DKK; Investigation: JK, SK; Visualization: JK, SK; Project administration: JK, CJL; Supervision: CJL, MC; Software: SK; Writing—original draft: JK, SK, DKK; Writing—review and editing: CJL, MC; JK and SK contributed equally to this study.
Corresponding authors
Ethics declarations
Conflict of interest
A patent was filed (KR 10–2023-0027219/2023.02.28) by the Institute for Basic Science (IBS).
Availability of code, data and materials
The data and code used in this study are available at the following GitHub repository: https://github.com/jeakwon/subtle. A demo site showcasing the application of the code is available at https://subtle.ibs.re.kr. Other materials, and codes used in the study are available upon request. All data and code are released under the GNU General Public License v3.0. The authors declare that the data and code support the findings of this study.
Additional information
Communicated by Anna Zamansky.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file 2 (mp4 17851 KB)
Supplementary file 3 (mp4 3916 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kwon, J., Kim, S., Kim, DK. et al. SUBTLE: An Unsupervised Platform with Temporal Link Embedding that Maps Animal Behavior. Int J Comput Vis (2024). https://doi.org/10.1007/s11263-024-02072-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11263-024-02072-0