Abstract
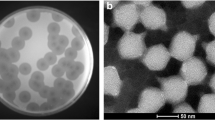
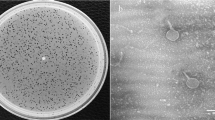
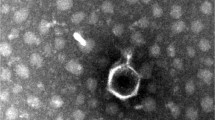
Phages are found in a wide variety of places where bacteria exist including body fluids. The aim of the present study was to isolate phages from the urine samples of patients with urinary tract infection. The 10 urine samples were cultured to isolate bacteria and also used as phage sources against the isolated bacteria. From 10 urine samples with positive cultures, 3 phages were isolated (33%) and two of them were further studied. The Klebsiella phage GADU21 and Escherichia phage GADU22 phages infected Klebsiella pneumonia and Escherichia coli, respectively. Among the tested 14 species for host range analysis, the Klebsiella phage GADU21 was able to infect two species which are Klebsiella pneumonia and Proteus mirabilis, and Escherichia phage GADU22 was able to infect four species which are Shigella flexneri, Shigella sonnei and Escherichia coli. Among different isolates of the indicator bacteria for each phage, GADU21 infected half of the tested 20 Klebsiella pneumonia isolates while GADU22 infected 85% of the tested 20 E. coli isolates. The genome sizes and GC ratios were 75,968 bp and 44.4%, and 168,023 bp and 35.3% for GADU21 and GADU22, respectively. GADU21 and GADU22 were both lytic and had no antibiotic resistance and virulence genes. GADU21 was homologue with Klebsiella phage vB_KpP_FBKp27 but only 88% of the genome was covered by this phage. The non-covered parts of the GADU21 genome included genes for tail-fiber-proteins and HNH-endonuclease. GADU22 had 94.8% homology with Escherichia phage vB_Eco_OMNI12 and had genes for immunity proteins. Phylogenetic analysis showed GADU21 and GADU22 were members of Schitoviridae family and Efbeekayvirus genus and Straboviridae family and Tevenvirinae genus, respectively. VIRIDIC analysis classified these phages in new species clusters. Our study demonstrated the possibility to use infected body fluids as phage sources to isolate novel phages. GADU21 is the first reported Klebsiella phage isolated from human body fluid. The absence of virulence and antibiotic resistance genes in their genomes makes the phages a potential therapeutic tool against infections.







Similar content being viewed by others
Data availability
The genome sequences of Klebsiella phage GADU21 and Escherichia phage GADU22 were deposited in GenBank under the accession number ON004963.1 and OQ944321, respectively.
References
D’Herelle F (2007) On an invisible microbe antagonistic toward dysenteric bacilli: brief note by Mr. F. D’Herelle, presented by Mr. Roux. 1917. Res Microbiol 158(7):553–554
d’Herelle F (1931) Phage as a treatment in acute medical and surgical infections. Bull N Y Acad Med 7(5):329–348
Żaczek M, Weber-Dąbrowska B, Międzybrodzki R, Łusiak-Szelachowska M, Górski A (2020) Phage therapy in Poland—a centennial journey to the first ethically approved treatment facility in Europe. Front Microbiol 11:1056
Aswani VH, Shukla SK (2021) An early history of phage therapy in the United States: is it time to reconsider? Clin Med Res 19(2):82–89
Barron M (2022). Phage therapy: past, present and future. https://asm.org/Articles/2022/August/Phage-Therapy-Past,-Present-and-Future
Patil A, Banerji R, Kanojiya P, Koratkar S, Saroj S (2021) Phages for ESKAPE: role in pathogenicity and measures of control. Expert Rev Anti Infect Ther 19(7):845–865
Denamur E, Clermont O, Bonacorsi S, Gordon D (2021) The population genetics of pathogenic Escherichia coli. Nat Rev Microbiol 19(1):37–54
Li Y, Kumar S, Zhang L, Wu H (2022) Klebsiella pneumonia and its antibiotic resistance: a bibliometric analysis. BioMed Res Int. https://doi.org/10.1155/2022/1668789
Hyman P (2019) Phages for phage therapy: isolation, characterization, and host range breadth. Pharmaceuticals 12(1):35
Karaynir A, Salih H, Bozdoğan B, Güçlü Ö, Keskin D (2022) Isolation and characterization of Brochothrix phage ADU4. Virus Res 321:198902
Pacífico C, Hilbert M, Sofka D, Dinhopl N, Pap IJ, Aspöck C et al (2019) Natural occurrence of Escherichia coli-infecting phages in clinical samples. Front Microbiol 10:2484
Ali S, Karaynir A, Salih H, Öncü S, Bozdoğan B (2023) Characterization, genome analysis and antibiofilm efficacy of lytic Proteus phages RP6 and RP7 isolated from university hospital sewage. Virus Res 326:199049
Salih H, Karaynir A, Yalcin M, Oryasin E, Holyavkin C, Basbulbul G, Bozdogan B (2022) Metagenomic analysis of wastewater phageome from a University Hospital in Turkey. Arch Microbiol 204(6):353
Arkin AP, Cottingham RW, Henry CS, Harris NL, Stevens RL, Maslov S et al (2018) KBase: the United States department of energy systems biology knowledgebase. Nat Biotechnol 36(7):566–569
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30(15):2114–2120
Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A (2020) Using SPAdes de novo assembler. Curr Protoc Bioinform 70(1):e102
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29(8):1072–1075
Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ et al (2015) RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5(1):8365
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36(suppl_2):W5–W9
Lowe TM, Chan PP (2016) tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44(W1):W54–W57
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinform 10:1–9
Wintersinger JA, Wasmuth JD (2015) Kablammo: an interactive, web-based BLAST results visualizer. Bioinformatics 31(8):1305–1306
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39(suppl_2):W29–W37
Hockenberry AJ, Wilke CO (2021) BACPHLIP: predicting bacteriophage lifestyle from conserved protein domains. PeerJ 9:e11396
Cosentino S, Voldby Larsen M, Møller Aarestrup F, Lund O (2013) PathogenFinder-distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE 8(10):e77302
Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Aarestrup FM (2020) ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrobial Chemother 75(12):3491–3500
Malberg Tetzschner AM, Johnson JR, Johnston BD, Lund O, Scheutz F (2020) In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J Clin Microbiol 58(10):10–1128
Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Stevens R (2020) The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res 48(D1):D606–D612
Boeckaerts D, Stock M, De Baets B, Briers Y (2022) Identification of phage receptor-binding protein sequences with hidden Markov models and an extreme gradient boosting classifier. Viruses 14(6):1329
Garneau JR, Depardieu F, Fortier LC, Bikard D, Monot M (2017) PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep 7(1):8292
Nishimura Y, Yoshida T, Kuronishi M, Uehara H, Ogata H, Goto S (2017) ViPTree: the viral proteomic tree server. Bioinformatics 33(15):2379–2380
UniProt Consortium (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47(D1):D506–D515
Navarro F, Muniesa M (2017) Phages in the human body. Front Microbiol 8:566
Sanmukh SG, Admella J, Moya-Andérico L, Fehér T, Arévalo-Jaimes BV, Blanco-Cabra N, Torrents E (2023) Accessing the in vivo efficiency of clinically isolated phages against uropathogenic and invasive biofilm-forming Escherichia coli strains for phage therapy. Cells 12(3):344
Brown-Jaque M, Muniesa M, Navarro F (2016) Phages in clinical samples can interfere with microbiological diagnostic tools. Sci Rep 6(1):1–8
Choi M, Hegerle N, Nkeze J, Sen S, Jamindar S, Nasrin S et al (2020) The diversity of lipopolysaccharide (O) and capsular polysaccharide (K) antigens of invasive Klebsiella pneumoniae in a multi-country collection. Front Microbiol 11:1249
de Sousa JA, Buffet A, Haudiquet M, Rocha EP, Rendueles O (2020) Modular prophage interactions driven by capsule serotype select for capsule loss under phage predation. ISME J 14(12):2980–2996
Palusiak A (2022) Proteus mirabilis and Klebsiella pneumoniae as pathogens capable of causing co-infections and exhibiting similarities in their virulence factors. Front Cell Infect Microbiol. https://doi.org/10.3389/fcimb.2022.991657
Kim SH, Adeyemi DE, Park MK (2021) Characterization of a new and efficient polyvalent phage infecting E. coli O157:H7 Salmonella spp., and Shigella sonnei. Microorganisms 9(10):2105. https://doi.org/10.3390/microorganisms9102105
Doore SM, Schrad JR, Dean WF, Dover JA, Parent KN (2018) Shigella phages isolated during a dysentery outbreak reveal uncommon structures and broad species diversity. J Virol 92(8):e02117-e2217
Lee H, Ku HJ, Lee DH, Kim YT, Shin H, Ryu S, Lee JH (2016) Characterization and genomic study of the novel phage HY01 infecting both Escherichia coli O157: H7 and Shigella flexneri: potential as a biocontrol agent in food. PLoS ONE 11(12):e0168985
Yang Q, Deng S, Xu J, Farooq U, Yang T, Chen W (2021) Poly (indole-5-carboxylic acid)/reduced graphene oxide/gold nanoparticles/phage-based electrochemical biosensor for highly specific detection of Yersinia pseudotuberculosis. Microchim Acta 188:1–13
Henning U, Jann K (1979) Two-component nature of phage T4 receptor activity in Escherichia coli K-12. J Bacteriol 137(1):664–666
Philipson CW, Voegtly LJ, Lueder MR, Long KA, Rice GK, Frey KG (2018) Characterizing phage genomes for therapeutic applications. Viruses 10(4):188
Liu Y, Mi Z, Mi L, Huang Y, Li P, Liu H (2019) Identification and characterization of capsule depolymerase Dpo48 from Acinetobacter baumannii phage IME200. PeerJ 7:e6173
Sun S, Gao S, Kondabagil K, Xiang Y, Rossmann MG, Rao VB (2012) Structure and function of the small terminase component of the DNA packaging machine in T4-like phages. Proc Natl Acad Sci 109(3):817–822
Lokareddy RK, Hou CFD, Li F, Yang R, Cingolani G (2022) Viral small terminase: a divergent structural framework for a conserved biological function. Viruses 14(10):2215
Kala S, Cumby N, Sadowski PD, Hyder BZ, Kanelis V, Davidson AR, Maxwell KL (2014) HNH proteins are a widespread component of phage DNA packaging machines. Proc Natl Acad Sci 111(16):6022–6027
Bailly-Bechet M, Vergassola M, Rocha E (2007) Causes for the intriguing presence of tRNAs in phages. Genome Res 17(10):1486–1495
Yang Z, Horton JR, Zhou L, Zhang XJ, Dong A, Zhang X (2003) Structure of the phage T4 DNA adenine methyltransferase. Nat Struct Mol Biol 10(10):849–855
Wang GR, Vianelli A, Goldberg EB (2000) Phage T4 self-assembly: in vitro reconstitution of recombinant GP2 into infectious phage. J Bacteriol 182(3):672–679
Hobbs SJ, Wein T, Lu A, Morehouse BR, Schnabel J, Leavitt A et al (2022) Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity. Nature 605(7910):522–526
Skorupski K, Tomaschewski J, Rüger W, Simon LD (1988) A phage T4 gene which functions to inhibit Escherichia coli Lon protease. J Bacteriol 170(7):3016–3024
Hsueh BY, Severin GB, Elg CA, Waldron EJ, Kant A, Wessel AJ (2022) Phage defence by deaminase-mediated depletion of deoxynucleotides in bacteria. Nat Microbiol 7(8):1210–1220
Abremski K, Black LW (1979) The function of phage T4 internal protein I in a restrictive strain of Escherichia coli. Virology 97(2):439–447
Kanamaru S, Uchida K, Nemoto M, Fraser A, Arisaka F, Leiman PG (2020) Structure and function of the T4 spackle protein Gp61. 3. Viruses 12(10):1070
Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Rüger W (2003) Phage T4 genome. Microbiol Mol Biol Rev 67(1):86–156
Korf IH, Meier-Kolthoff JP, Adriaenssens EM, Kropinski AM, Nimtz M, Rohde M (2019) Still something to discover: novel insights into Escherichia coli phage diversity and taxonomy. Viruses 11(5):454
Fabijan AP, Kamruzzaman M, Martinez-Martin D, Venturini C, Mickiewicz K, Flores-Rodriguez N (2021) L-form switching confers antibiotic, phage and stress tolerance in pathogenic Escherichia coli. biorxiv. https://doi.org/10.1101/2021.06.21.449206
Fokine A, Islam MZ, Zhang Z, Bowman VD, Rao VB, Rossmann MG (2011) Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like phage. J Virol 85(16):8141–8148
Murphy J, Klumpp J, Mahony J, O’Connell-Motherway M, Nauta A, van Sinderen D (2014) Methyltransferases acquired by lactococcal 936-type phage provide protection against restriction endonuclease activity. BMC Genomics 15(1):1–11
Mijbel Ali B, Gatea Kaabi SA, Al-Bayati MA, Musafer HK (2021) A novel phage cocktail therapy of the urinary tract infection in a mouse model. Arch Razi Inst 76(5):1229–1236
Le T, Nang SC, Zhao J, Yu HH, Li J, Gill JJ (2023) Therapeutic potential of intravenous phage as standalone therapy for recurrent drug-resistant urinary tract infections. Antimicrobial Agents Chemother 67(4):e00037-e42
Acknowledgements
The author, Hanife Salih Doğan, is supported by YOK 100/2000 fellowship program during the PhD program and 2211/A National PhD Scholarship Program, The Scientific and Technological Research Council of Turkey.
Funding
No funding was received for this research/project.
Author information
Authors and Affiliations
Contributions
HSD: Data curation, investigation, methodology, writing—original draft, writing—review and editing, AK: Conceptualization, investigation, methodology, ÜY: Methodology, resources, BB: Methodology, resources, TH: Conceptualization, resources, supervision, writing—review and editing, BB: Conceptualization, funding acquisition, project administration, supervision, writing—review and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This study is approved under the ethical approval code of Ethical Committee of Health Sciences University Gülhane Training and Research Hospital IC-2020/315. Informed consent from the participants was obtained.
Additional information
Edited by Andrew Millard.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11262_2024_2052_MOESM1_ESM.jpg
Figure S1. VipTree analysis of Klebsiella phage GADU21 and Escherichia phage GADU22 with reference dsDNA viruses (A) GADU21 and GADU22 phages were represented with stars. The outer ring indicates hosts of the phages, and the inner ring indicates the family to which the phages belong. The hosts of these phages were founded to belong to Gammaproteobacteria (Pseudomonata) phylum. (B) The taxonomic relationship for GADU21 and GADU22 phages and their relatives showed that GADU21 phage is a member of Schitoviridae family and Efbeekayvirus genus and GADU22 phage is a member of Straboviridae family and Tevenvirinae genus. Supplementary file1 (JPG 290 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Salih Doğan, H., Karaynir, A., Yilmaz, Ü.İ. et al. Two novel phages, Klebsiella phage GADU21 and Escherichia phage GADU22, from the urine samples of patients with urinary tract infection. Virus Genes 60, 208–221 (2024). https://doi.org/10.1007/s11262-024-02052-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-024-02052-z