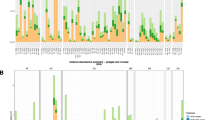
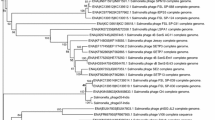
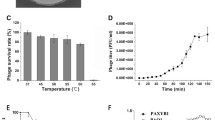
Abstract
Phage DNA analysis gives opportunity to understand living ecosystem of the environment where the samples are taken. In the present study, we analyzed phage DNA obtained from wastewater sample of university hospital sewage. After filtration, long high-speed centrifugation was done to collect phages. DNA was extracted from pellet by phenol chloroform extraction and used for NGS sequencing. The host profile, taxonomic and functional analyses were performed using MG-RAST, and ResFinder program was used for resistance gene detection. High amounts of reads belong to bacteriophage groups (~ 95%) from our DNA sample were obtained and all bacteriophage reads were found belonging to Caudovirales order and Myoviridae (56%), Siphoviridae (43%), and Podoviridae (0.02%) families. The most common host genera were Escherichia (88.20%), Salmonella (5.49%) and Staphylococcus (5.19%). SEED subsystems hits were mostly structural parts and KEGG Orthology hits were nucleotide- and carbohydrate metabolism-related genes. No anti-microbial resistance genes were detected. Our bacteriophage DNA purification method is favorable for phage metagenomic studies. Dominance of coliphages may explain infrequent Podoviridae. Dominancy of structural genes and auxiliary genes is probably due to abundance of lytic phages in our sample. Absence of antibiotic resistance genes even in hospital environment phages indicates that phages are not important carrier of resistance genes.






Similar content being viewed by others
References
Ackermann HW (2011) Bacteriophage taxonomy. Microbiol Aust 32(2):90–94
Adriaenssens EM, Sullivan MB, Knezevic P, van Zyl LJ, Sarkar BL, Dutilh BE et al (2020) Taxonomy of prokaryotic viruses: 2018–2019 update from the ICTV bacterial and archaeal viruses subcommittee. Arch Virol 165(5):1253–1260. https://doi.org/10.1007/s00705-020-04577-8
Arndt D, Grant J, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS (2016) PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44(W1):W16–W21. https://doi.org/10.1093/nar/gkw387
Balcazar JL (2014) Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLoS Pathog 10(7):e1004219
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Borrego JJ, Moriñigo MA, de Vicente A, Córnax R, Romero P (1987) Coliphages as an indicator of faecal pollution in water. Its relationship with indicator and pathogenic microorganisms. Water Res 21(12):1473–1480. https://doi.org/10.1016/0043-1354(87)90130-8
Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V et al (2020) ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75(12):3491–3500. https://doi.org/10.1093/jac/dkaa345
Breitbart M, Bonnain C, Malki K, Sawaya NA (2018) Phage puppet masters of the marine microbial realm. Nat Microbiol 3(7):754–766. https://doi.org/10.1038/s41564-018-0166-y
Cantalupo PG, Calgua B, Zhao G, Hundesa A, Wier AD, Katz JP et al (2011) Raw sewage harbors diverse viral populations. Mbio 2(5):e00180-e211. https://doi.org/10.1128/mBio.00180-11
Chaturongakul S, Ounjai P (2014) Phage–host interplay: examples from tailed phages and Gram-negative bacterial pathogens. Front Microbiol 5:442. https://doi.org/10.3389/fmicb.2014.00442
Chen Y, Wang Y, Paez-Espino D, Polz MF, Zhang T (2021) Prokaryotic viruses impact functional microorganisms in nutrient removal and carbon cycle in wastewater treatment plants. Nat Commun 12(1):1–11. https://doi.org/10.1038/s41467-021-25678-1
Chopyk J, Nasko DJ, Allard S, Bui A, Pop M, Mongodin EF, Sapkota AR (2020a) Seasonal dynamics in taxonomy and function within bacterial and viral metagenomic assemblages recovered from a freshwater agricultural pond. Environ Microbiome 15(1):1–16. https://doi.org/10.1186/s40793-020-00365-8
Chopyk J, Nasko DJ, Allard S, Callahan MT, Bui A, Ferelli AMC et al (2020b) Metagenomic analysis of bacterial and viral assemblages from a freshwater creek and irrigated field reveals temporal and spatial dynamics. Sci Total Environ 706:135395. https://doi.org/10.1016/j.scitotenv.2019.135395
Clokie MR, Kropinski AM, Lavigne R (2009) Bacteriophages: methods and protocols, volume 2: molecular and applied aspects. Humana Press, Leicester, pp 3–46
Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GG, Boling L et al (2014) A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun 5(1):1–11. https://doi.org/10.1038/ncomms5498
Dwivedi B, Xue B, Lundin D, Edwards RA, Breitbart M (2013) A bioinformatic analysis of ribonucleotide reductase genes in phage genomes and metagenomes. BMC Evol Biol 13(1):1–17. https://doi.org/10.1186/1471-2148-13-33
Enault F, Briet A, Bouteille L, Roux S, Sullivan MB, Petit MA (2017) Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J 11(1):237–247. https://doi.org/10.1038/ismej.2016.90
Enav H, Mandel-Gutfreund Y, Béjà O (2014) Comparative metagenomic analyses reveal viral-induced shifts of host metabolism towards nucleotide biosynthesis. Microbiome 2(1):1–12. https://doi.org/10.1186/2049-2618-2-9
García-Aljaro C, Ballesté E, Muniesa M, Jofre J (2017) Determination of crAssphage in water samples and applicability for tracking human faecal pollution. Microb Biotechnol 10(6):1775–1780. https://doi.org/10.1111/1751-7915.12841
Gulino K, Rahman J, Badri M, Morton J, Bonneau R, Ghedin E (2020) Initial mapping of the New York City wastewater virome. mSystems 5(3):e00876-19. https://doi.org/10.1128/mSystems.00876-19
Hayes S, Mahony J, Nauta A, Van Sinderen D (2017) Metagenomic approaches to assess bacteriophages in various environmental niches. Viruses 9(6):127. https://doi.org/10.3390/v9060127
Honap TP, Sankaranarayanan K, Schnorr SL, Ozga AT, Warinner C, Lewis CM Jr (2020) Biogeographic study of human gut-associated crAssphage suggests impacts from industrialization and recent expansion. PLoS ONE 15(1):e0226930. https://doi.org/10.1371/journal.pone.0226930
Hubeny J, Harnisz M, Korzeniewska E, Buta M, Zieliński W, Rolbiecki D et al (2021) Industrialization as a source of heavy metals and antibiotics which can enhance the antibiotic resistance in wastewater, sewage sludge and river water. PLoS ONE 16(6):e0252691. https://doi.org/10.1371/journal.pone.0252691
Jain R, Srivastava R (2009) Metabolic investigation of host/pathogen interaction using MS2-infected Escherichia coli. BMC Syst Biol 3(1):1–11. https://doi.org/10.1186/1752-0509-3-121
Jin M, Guo X, Zhang R, Qu W, Gao B, Zeng R (2019) Diversities and potential biogeochemical impacts of mangrove soil viruses. Microbiome 7(1):1–15. https://doi.org/10.1186/s40168-019-0675-9
Jofre J, Bosch A, Lucena F, Girones R, Tartera C (1986) Evaluation of Bacteroides fragilis bacteriophages as indicators of the virological quality of water. Water Sci Technol 18(10):167–173. https://doi.org/10.2166/wst.1986.0126
Jofre J, Lucena F, Blanch AR, Muniesa M (2016) Coliphages as model organisms in the characterization and management of water resources. Water 8(5):199. https://doi.org/10.3390/w8050199
Kasman LM, Porter LD (2021) Bacteriophages. In: StatPearls StatPearls Publishing. https://www.ncbi.nlm.nih.gov/books/NBK493185/
Keegan KP, Glass EM, Meyer F (2016) MG-RAST, a metagenomics service for analysis of microbial community structure and function. In: Microbial environmental genomics (MEG) (pp 207–233). Humana Press, New York. https://doi.org/10.1007/978-1-4939-3369-3_13
Korf IH, Meier-Kolthoff JP, Adriaenssens EM, Kropinski AM, Nimtz M, Rohde M et al (2019) Still something to discover: novel insights into Escherichia coli phage diversity and taxonomy. Viruses 11(5):454. https://doi.org/10.3390/v11050454
Kumar N, Gupta AK, Sudan SK, Pal D, Randhawa V, Sahni G et al (2021) Abundance and diversity of phages, microbial taxa, and antibiotic resistance genes in the sediments of the river ganges through metagenomic approach. Microb Drug Resist. https://doi.org/10.1089/mdr.2020.0431
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Meth 9(4):357–359. https://doi.org/10.1038/nmeth.1923
Lindell D, Sullivan MB, Johnson ZI, Tolonen AC, Rohwer F, Chisholm SW (2004) Photosynthesis genes in Prochlorococcus cyanophage. Proc Natl Acad Sci USA 101:3–8
McMinn BR, Ashbolt NJ, Korajkic A (2017) Bacteriophages as indicators of faecal pollution and enteric virus removal. Lett Appl Microbiol 65(1):11–26. https://doi.org/10.1111/lam.12736
McNair K, Bailey BA, Edwards RA (2012) PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 28(5):614–618. https://doi.org/10.1093/bioinformatics/bts014
Méndez J, Toribio-Avedillo D, Mangas-Casas R, Martínez-González J (2020) Bluephage, a method for efficient detection of somatic coliphages in one hundred milliliter water samples. Sci Rep 10(1):1–5. https://doi.org/10.1038/s41598-020-60071-w
Muniesa M, Lucena F, Jofre J (1999) Study of the potential relationship between the morphology of infectious somatic coliphages and their persistence in the environment. J Appl Microbiol 87(3):402–409. https://doi.org/10.1046/j.1365-2672.1999.00833.x
Muniesa M, Lucena F, Blanch AR, Payán A, Jofre J (2012) Use of abundance ratios of somatic coliphages and bacteriophages of Bacteroides thetaiotaomicron GA17 for microbial source identification. Water Res 46(19):6410–6418. https://doi.org/10.1016/j.watres.2012.09.015
Nayfach S, Páez-Espino D, Call L, Low SJ, Sberro H, Ivanova NN et al (2021) Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat Microbiol 6(7):960–970. https://doi.org/10.1038/s41564-021-00928-6
Neelakanta G, Sultana H (2013) The use of metagenomic approaches to analyze changes in microbial communities. Microbiol Insights 6:S10819. https://doi.org/10.4137/MBI.S10819
Owen SV, Canals R, Wenner N, Hammarlöf DL, Kröger C, Hinton JC (2020) A window into lysogeny: revealing temperate phage biology with transcriptomics. Microb Genom 6:2. https://doi.org/10.1099/mgen.0.000330
Petrovich ML, Ben Maamar S, Hartmann EM, Murphy BT, Poretsky RS, Wells GF (2019) Viral composition and context in metagenomes from biofilm and suspended growth municipal wastewater treatment plants. Microb Biotechnol 12(6):1324–1336. https://doi.org/10.1111/1751-7915.13464
Petrovich ML, Zilberman A, Kaplan A, Eliraz GR, Wang Y, Langenfeld K et al (2020) Microbial and viral communities and their antibiotic resistance genes throughout a hospital wastewater treatment system. Front Microbiol 11:153. https://doi.org/10.3389/fmicb.2020.00153
Potapov SA, Tikhonova IV, Krasnopeev AY, Kabilov MR, Tupikin AE, Chebunina NS et al (2019) Metagenomic analysis of virioplankton from the pelagic zone of Lake Baikal. Viruses 11(11):991. https://doi.org/10.3390/v11110991
Ramos-Vivas J, Elexpuru-Zabaleta M, Samano ML, Barrera AP, Forbes-Hernández TY, Giampieri F, Battino M (2021) Phages and enzybiotics in food biopreservation. Molecules 26(17):5138
Reis-Cunha JL, Bartholomeu DC, Manson AL, Earl AM, Cerqueira GC (2019) ProphET, prophage estimation tool: a stand-alone prophage sequence prediction tool with self-updating reference database. PLoS ONE 14(10):e0223364. https://doi.org/10.1371/journal.pone.0223364
Rezaeinejad S, Vergara GGRV, Woo CH, Lim TT, Sobsey MD, Gin KYH (2014) Surveillance of enteric viruses and coliphages in a tropical urban catchment. Water Res 58:122–131
Rohwer F, Segall A, Steward G, Seguritan V, Breitbart M, Wolven F, Farooq Azam F (2000) The complete genomic sequence of the marine phage Roseophage SIO1 shares homology with nonmarine phages. Limnol Oceanogr 45(2):408–418. https://doi.org/10.1016/j.watres.2014.03.051
Roux S, Krupovic M, Debroas D, Forterre P, Enault F (2013) Assessment of viral community functional potential from viral metagenomes may be hampered by contamination with cellular sequences. Open Biol 3(12):130160. https://doi.org/10.1098/rsob.130160
Rowe WP, Baker-Austin C, Verner-Jeffreys DW, Ryan JJ, Micallef C, Maskell DJ, Pearce GP (2017) Overexpression of antibiotic resistance genes in hospital effluents over time. J Antimicrob Chemother 72(6):1617–1623
Sambrook J, Russell DW (2006) Purification of nucleic acids by extraction with phenol: chloroform. Cold Spring Harb Protoc 2006:prot4455
Sharma A, Schmidt M, Kiesel B, Mahato NK, Cralle L, Singh Y et al (2018) Bacterial and archaeal viruses of Himalayan hot springs at Manikaran modulate host genomes. Front Microbiol 9:3095. https://doi.org/10.3389/fmicb.2018.03095
Shkoporov AN, Hill C (2019) Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe 25(2):195–209. https://doi.org/10.1016/j.chom.2019.01.017
Skvortsov T, De Leeuwe C, Quinn JP, McGrath JW, Allen CC, McElarney Y et al (2016) Metagenomic characterisation of the viral community of Lough Neagh, the largest freshwater lake in Ireland. PLoS ONE 11(2):e0150361. https://doi.org/10.1371/journal.pone.0150361
Sørensen PE, Van Den Broeck W, Kiil K, Jasinskyte D, Moodley A, Garmyn A et al (2020) new insights into the biodiversity of coliphages in the intestine of poultry. Sci Rep 10(1):1–12. https://doi.org/10.1038/s41598-020-72177-2
Ssekatawa K, Byarugaba DK, Kato CD, Wampande EM, Ejobi F, Tweyongyere R, Nakavuma JL (2021) A review of phage mediated antibacterial applications. Alexandria J Med 57(1):1–20
Stachler E, Bibby K (2014) Metagenomic evaluation of the highly abundant human gut bacteriophage CrAssphage for source tracking of human fecal pollution. Environ Sci Technol Lett 1(10):405–409. https://doi.org/10.1021/ez500266s
Stachler E, Kelty C, Sivaganesan M, Li X, Bibby K, Shanks OC (2017) Quantitative CrAssphage PCR assays for human fecal pollution measurement. Environ Sci Technol 51(16):9146–9154. https://doi.org/10.1021/acs.est.7b02703
Subirats J, Sànchez-Melsió A, Borrego CM, Balcázar JL, Simonet P (2016) Metagenomic analysis reveals that bacteriophages are reservoirs of antibiotic resistance genes. Int J Antimicrob Agents 48(2):163–167. https://doi.org/10.1016/j.ijantimicag.2016.04.028
Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, Chisholm SW (2011) Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. PNAS 108(39):E757–E764. https://doi.org/10.1073/pnas.1102164108
Toribio-Avedillo D, Blanch AR, Muniesa M, Rodríguez-Rubio L (2021) Bacteriophages as fecal pollution indicators. Viruses 13(6):1089. https://doi.org/10.3390/v13061089
Turner D, Kropinski AM, Adriaenssens EM (2021) A roadmap for genome-based phage taxonomy. Viruses 13(3):506. https://doi.org/10.3390/v13030506
UniProt (2021) The universal protein knowledgebase in 2021. Nucleic Acids Research 49D1:D480–D489. https://www.uniprot.org/uniprot/A0A023ZX80
Wang M, Xiong W, Liu P, Xie X, Zeng J, Sun Y, Zeng Z (2018) Metagenomic insights into the contribution of phages to antibiotic resistance in water samples related to swine feedlot wastewater treatment. Front Microbiol 9:2474. https://doi.org/10.3389/fmicb.2018.02474
Wikner J, Vallino JJ, Steward GF, Smith DC, Azam F (1993) Nucleic acids from the host bacterium as a major source of nucleotides for three marine bacteriophages. FEMS Microbiol Ecol 12(4):237–248. https://doi.org/10.1111/j.1574-6941.1993.tb00036.x
Yang S, Gao X, Meng J, Zhang A, Zhou Y, Long M et al (2018) Metagenomic analysis of bacteria, fungi, bacteriophages, and helminths in the gut of giant pandas. Front Microbiol 9:1717. https://doi.org/10.3389/fmicb.2018.01717
Yasir M (2021) Analysis of microbial communities and pathogen detection in domestic sewage using metagenomic sequencing. Divers 13(1):6. https://doi.org/10.3390/d13010006
Zechner V, Sofka D, Paulsen P, Hilbert F (2020) Antimicrobial resistance in Escherichia coli and resistance genes in coliphages from a small animal clinic and in a patient dog with chronic urinary tract infection. Antibiotics 9(10):652. https://doi.org/10.3390/antibiotics9100652
Zhen X, Lundborg CS, Sun X, Hu X, Dong H (2019) Economic burden of antibiotic resistance in ESKAPE organisms: a systematic review. Antimicrob Resist Infect Control 8(1):1–23
Acknowledgements
The authors, Hanife Salih and Melis Yalcin, are supported by YOK 100/2000 fellowship program during the PhD.
Funding
No funding was received for conducting this study.
Author information
Authors and Affiliations
Contributions
HS: methodology, formal analysis, investigation, writing—original draft, visualization; AK: methodology, investigation, resources; MY: methodology, investigation; EO: resources, writing—review & editing; CH: formal analysis; GB: writing—review & editing, supervision; BB: conceptualization, writing—review & editing, visualization, supervision, project administration. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interests
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Additional information
Communicated by Ran Wang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Salih, H., Karaynir, A., Yalcin, M. et al. Metagenomic analysis of wastewater phageome from a University Hospital in Turkey. Arch Microbiol 204, 353 (2022). https://doi.org/10.1007/s00203-022-02962-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-02962-2