Abstract
Chikungunya virus (CHIKV), a (re)emerging arbovirus, is the causative agent of chikungunya fever. To date, no approved vaccine or specific antiviral therapy are available. CHIKV has repeatedly been responsible for serious economic and public health impacts in countries where CHIKV epidemics occurred. Antiviral tests in vitro are generally performed in Vero-B4 cells, a well characterised cell line derived from the kidney of an African green monkey. In this work we characterised a CHIKV patient isolate from Brazil (CHIKVBrazil) with regard to cell affinity, infectivity, propagation and cell damage and compared it with a high-passage lab strain (CHIKVRoss). Infecting various cell lines (Vero-B4, A549, Huh-7, DBTRG, U251, and U138) with both virus strains, we found distinct differences between the two viruses. CHIKVBrazil does not cause cytopathic effects (CPE) in the human hepatocarcinoma cell line Huh-7. Neither CHIKVBrazil nor CHIKVRoss caused CPE on A549 human lung epithelial cells. The human astrocyte derived glioblastoma cell lines U138 and U251 were found to be effective models for lytic infection with both virus strains and we discuss their predictive potential for neurogenic CHIKV disease. We also detected significant differences in antiviral efficacies regarding the two CHIKV strains. Generally, the antivirals ribavirin, hydroxychloroquine (HCQ) and T-1105 seem to work better against CHIKVBrazil in glioblastoma cells than in Vero-B4. Finally, full genome analyses of the CHIKV isolates were done in order to determine their lineage and possibly explain differences in tissue range and antiviral compound efficacies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Taxonomy, structure, genome organisation, ecology, and epidemiology
Chikungunya virus (CHIKV) is an arthropod borne (arbo-) virus of the alphavirus genus. Belonging to the “Old World” viruses, CHIKV is categorised as an arthritogenic alphavirus due to the primary site of disease manifestation, the joints [1].
To date three CHIKV phylogroups and one distinct sublineage are known. The phylogroups consist of the West African (WA), East-Central-South African (ECSA) and Asian genotype [2]. The Indian Ocean Outbreak, which started in Kenya in 2004, was caused by a mutated sublineage that is referred to as the Indian Ocean Lineage (IOL) and originated from of the ECSA isolates [3].
CHIKV is an enveloped virus and the virion contains single-standed, positive-sense RNA of about 11,800 nucleotides [4]. The virus has the general structure of all alphaviruses (for details on structure, epidemiology, and pathogenesis see Hucke, Bestehorn-Willmann [5] and Hucke and Bugert [6]).
CHIKV is generally transmitted to humans by the bite of an infected mosquito from the Aedes family, mainly Aedes aegypti and Aedes albopictus [7]. After entering the skin, viral replication and amplification seem to occur mainly in dermal fibroblasts [8]. Dendritic cells capture virus particles, transport them to the nearest lymph nodes where blood monocytes and macrophages are infected. At this point viremia sets in [9]. Via blood stream CHIKV then reaches the muscles and joints. Infection of these sites causes the main symptoms of CHIKF—myalgia and arthralgia. Infection of the joints often results in cartilage degradation and bone loss [10], which explains the severe and debilitating arthralgia that are the hallmark of the disease and gave the virus its name. After the acute phase of the illness has passed, myalgia and arthralgia can go into a chronic state and last for months or even years, leaving the patient with a severely deteriorated quality of life.
Apart from these well-known sites of infection, CHIKV has been known to infect a wide range of secondary organs which may cause severe complications in patients [7]. Although CHIKV has originally not been classified as a neurotropic virus, the La Reunion outbreak recorded an increased number of neurological complications (e.g. meningitis, encephalitis, febrile seizures, Guillain Barré syndrome, neuro-ocular diseases), especially in the elderly and the very young [11, 12]. It was demonstrated that CHIKV is able to replicate in neurons, astrocytes, oligodendrocytes, and microglia cells [13].
To date, no approved vaccine or specific antiviral therapies are available. Considering the time it takes to fully recover from CHIKV disease, an effective antiviral is of utmost importance. A variety of antivirals curb CHIKV infection in vitro but lack efficacy in vivo [6]. Well established antivirals for in vitro assays are chloroquine/hydroxychloroquine, ribavirin, and favipiravir, although they show significant differences in their efficacy depending on the virus strain and cell line [6].
So far, little focus has been given on which human cell lines are suitable for in vitro studies with CHIKV. Also the question on whether different virus strains show different cell affinities in relevant human cell lines has not been addressed properly. Furthermore, antivirals might have different efficacies depending on the cell line and the virus strain. There is the possibility that high-passage, laboratory-adapted strains (such as CHIKVRoss) are able to replicate in cell lines which are not affected by wt CHIKV infection. This raises the question to which extent such high-passage reference strains are still comparable to field strains in regard of antiviral efficacies.
To shed light on these questions, two different CHIKV strains, the high-passage Ross strain, isolated in 1953 (CHIKVRoss), and a field isolate from Brazil, isolated in 2015 (CHIKVBrazil) were compared with regard to cell affinity and drug sensitivity towards well established antiviral substances. Finally, a whole genome sequence comparison of both strains was performed to try to explain differences in cell affinity or drug sensitivities on a genomic level.
Materials and methods
Cells and cell culture
Vero-B4 cells (ATCC® CCL-81™) [14], A549 cells (ATCC® CCL-185™) [15], Huh-7 cells (JCRB0403) [16], the glioblastoma cell line DBTRG-05MG (ATCC® CRL-2020™) [17], were obtained from ATCC whilst the human glioblastoma cell lines U138 (aka U-138 MG, ATCC® HTB-16™) and U251 (aka U-251 MG, ATCC® HTB-17™; formerly known as U-373 MG) were a gift of R. Brack-Werner, Institute of Virology, German Research Center for Environmental Health (GmbH).
Dulbecco’s Modified Eagle Medium (DMEM(1X) + GlutaMAX™-I medium, Thermo Fisher Scientific Ltd, UK), with either 1 g/L of D-glucose (in the following referred to as “Low Glucose” (LG)) or with 4.5 g/L of D-glucose (“High Glucose” (HG)) were used. 5% heat inactivated foetal bovine serum (FBS; Sigma-Aldrich, Hilden, Germany) was added. U138 and U251 cells were kept on DMEM HG medium whilst Vero-B4, A549, DBTRG, and Huh-7 were kept on DMEM LG.
Antiviral substances
The antiviral compound T-1105 was provided by the School of Pharmacy and Pharmaceutical Sciences of the Cardiff University, UK. T-1105 is a direct nucleoside (purine) analogue and the defluorinated analogue of favipiravir (T-705). The compound was provided as a solid powder and was dissolved in DMSO to create a 10 mM solution.
Other antiviral substances used as controls were ribavirin (RBV), and hydroxychloroquine (HCQ) (both from Sigma-Aldrich). RBV and HCQ were dissolved in purified water to create stock solutions of 100 mM and 10 mM, respectively. For further dilutions DMEM LG was used.
Virus
Viruses used in this study are part of the BSL3 reference collection of the Bundeswehr Institute of Microbiology (IMB), Munich. The wildtype CHIKV strain L3-4497 originates from a patient isolate from Brazil (CHIKVBrazil; 2015). Subpassaged samples of the initial cultivation (Vero-B4) were used to establish a working stock of CHIKV (also grown on Vero-B4). In this study the wildtype CHIKV strain used had previously been passaged twice on Vero-B4 cells after its isolation. GenBank accession number: Banklt2561907 Chikungunya_Brazil_4497 ON009842.
The lab attenuated CHIKV Ross strain L3-3950 (CHIKVRoss; NH177) has been isolated from an outbreak in Tanzania in 1953 [18,19,20]. GenBank accession number: Banklt2561907 Chikungunya_Ross_NH177 ON009843.
Both virus strains belong to the ESCA genotype.
Virus stock production
Vero-B4 cells were cultivated in a T75 flask in DMEM LG with 5% FBS until they reached 80% confluence. After removal of supernatant and a one-time washing with DMEM LG, 500 µL of the original virus stock suspension from the L3 reference stocks were added to the T75 cell culture flask and canted gently to ensure the virus reached the entire cell layer. After one minute, 20 mL of DMEM LG with 5% FBS were added to the bottle and subsequently flasks were incubated at 37 °C and 5% CO2 until maximal cytopathic effect (CPE) was observed via microscope (Zeiss Axiovert25, Germany).
Two to three days post infection the supernatant of the bottle was collected, FBS was added to a final concentration of 20%, and the virus solution was aliquoted into 1 mL cryotubes with 500 µL of CHIKV suspension each and stored at − 70 °C. Virus stock titres were evaluated via plaque assay.
Virus tittering via plaque assay
One mL of Vero-B4 and U138 cells (1.2 × 105 cells/mL) were seeded into a 24-well plate and allowed to settle overnight. The next day, the supernatant of the cells was removed and cells were infected with 200 µL of a tenfold serial dilution (DMEM LG) of CHIKVBrazil or CHIKVRoss (Vero-B4 cells only).
The plate was gently swayed and incubated for 30 min at 37 °C and 5% CO2.
Then, 800 µL of 0.8–1% carboxymethylcellulose (CMC) (Sigma-Aldrich, Schnelldorf, Germany) dissolved in MilliQ water, sterilised by autoclaving, mixed with DMEM and 2.5% FBS, was carefully added to each well using a multipette (Eppendorf, Germany). The plate was then incubated at 37 °C with 5% CO2 and observed daily for CPE with a microscope. Three to four days pi the cells were fixed and dyed by adding 1 mL of crystal violet (aqueous solution with 0.2% certified crystal violet and 20% formaldehyde (both from Merck, Darmstadt, Germany)) directly to each well. The plate was then incubated in the fridge at 4 °C overnight. Plates were then gently washed with distilled water until all the CMC and superfluous dye had been removed. Plaque assays with Vero-B4 cells were repeated at least 3 times independently. Assays with U138 cells were repeated twice.
Cell viability assay with MTS and data evaluation
Unless stated otherwise, cells were seeded at a density of 1 × 104 cells/100 µL/well in DMEM with 5% FBS in clear 96-well plates and allowed to settle overnight. The plates were incubated at 37 °C with 5% CO2 and 95–99% relative humidity. For treatment 50 µL of compound dilution were added to the corresponding wells. Virus infection was done with 50 µL of CHIKV dilution one hour after treatment. Toxicity assays and untreated non-infected (Mock) control were done adding 50 µL of medium instead of virus dilution. Final FBS concentration in the treated/infected wells was 2.5%. The plates were then incubated for 4 days.
All cell viability assays were done using the CellTiter 96®AQueous Non-Radioactive Cell Proliferation Assay (MTS) (Promega, USA) according to the manufacturer’s protocol with the difference that 20 µL MTS solution were used per 200 µL of experimental volume. Absorbance was measured at 490 nm with a reference wavelength of 620 nm using an ELISA plate reader (iMark™ Mikroplate Reader).
Apart from IC50/CC50 evaluation, the Optical Density (OD) values obtained were put into relation to Mock control with Microsoft Excel. Mock thus represents 100% viable cells in the column graphs. All graphs were prepared using GraphPad Prism 6 Software.
For comparisons of the different virus strains, ordinary one-way ANOVA tests were done (GraphPad). Probabilities of the test results are given with p-values.
Raw data values were put into relation with Mock control (Mock = 100%) and the positive control (untreated infected cells = 0%) in Excel. For calculation of IC50 and CC50 values, a dose–response curves equation (using raw data) of GraphPad Prism 6 was applied. The programme then calculated the relative IC50 value in relation to the raw data values of the most efficient compound concentration. Goodness of fit and plausible range are given by R2 and 95% Confidence Interval (95% CI). If a raw data value deviated more than 20% from the mean of the repeats, this particular value was omitted.
Kill curves
Apart from Huh-7 cells, all cells were seeded at a density of 1 × 104 cells/100 µl/well in DMEM with 5% FBS in 96-well plates. Huh-7 cells were seeded with only 5 × 103 cells/100 µl/well, due non-linear readout with CellTiter 96®AQueous Non-Radioactive Cell Proliferation Assay at higher concentration. After settling overnight, the cells were infected with 50 µL of virus dilutions ranging from 0 to 10–5 and incubated for 30 min. Then 50 µL of DMEM were added. Kill curve infection experiments were repeated at least thrice independently, with three technical replicates. Cell viability was evaluated using MTS.
Comparison of compound efficacy
RBV, HCQ, and T-1105 were used in concentrations previously published to inhibit wt CHIKV in Vero cells [21, 22]. The concentration used in our experiments were thus: RBV at 410 µM, HCQ at 10 µM, T-1105 at 10 µM and 50 µM.
As T-1105 was dissolved in DMSO, final DMSO concentration in all wells of the assay was uniformly 0.1% (Mock and positive control as well) to make sure the controls were unbiased by the solvent.
Treatment and infection of the cells were done as described in the IC50/CC50 experiments with the difference that multiplicity of infection (MOI) was 0.64. Each compound concentration had three or six technical replicates and the experiments were repeated at least thrice independently.
IC50/CC50 evaluation of RDV, HCQ, and T-1105 in Vero-B4 and U138 cells
For IC50/CC50 evaluation Vero-B4 and U138 cells were used. Serial dilutions of the compounds (RBV, HCQ, and T-1105) were prepared in assay medium (DMEM LG). To avoid precipitation of T-1105, a final concentration of 0.3% DMSO was kept in all wells containing this compound (and in the corresponding control wells). Serial dilutions of RBV ranged from 10 to 500 µM in U138 cells and 200 µM to 1000 µM in Vero-B4 and the toxicity assays. Serial dilutions of HCQ and T-1105 ranged from 1 to 100 µM. A volume of 50 µL of the compound dilution was added to the cells. Infection was done at a MOI of 0.355 with the CHIKV strain Brazil. As T-1105 had DMSO as a supplement to ensure solubility, two different kind of Mock and positive control (untreated infected cells) were run along, one with 0.3% of DMSO and the other without. Each compound was repeated at least thrice independently with three technical replicates.
Whole genome sequencing of chikungunya virus L3-4497 strain Brazil and Ross L3-3950 from InstMikroBio BW
For sequencing one vial of the respective stock solutions of CHIKVBrazil/Ross was used and the total RNA was purified using the Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manual. For Library preparation the NEBNext® Ultra™ II RNA Library Prep Kit for Illumina® was used according to the manufacturer’s protocol. Paired-end sSequencing of the generated libraries was performed on an Illumina MiSeq platform using a Miseq Reagent Kit V2 500 cycles chemistry.
De novo assemblies were generated for the two samples using the tool SPAdes version: 3.14.1. Pairwise alignments of the two generated whole genomes were generated using the ClustalW algorithm.
Results
Genome differences between the two virus strains
The CHIKV virus strains CHIKVRoss and CHIKVBrazil belong to the ECSA genotype. Genome analysis revealed 57 amino acid differences in the structural and non-structural polyproteins between our Brazilian field isolate and the Ross strain as shown in Fig. 1. For complete genome sequences of both virus strains see GenBank accession numbers Banklt2561907 Chikungunya_Ross_NH177 ON009843 and Banklt2561907 Chikungunya_Brazil_4497 ON009842.
Kill curve experiments
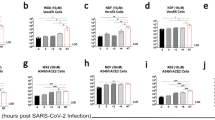
Vero-B4 cells are very sensitive to CHIKV infection. Even at an MOI of 0.000645 CHIKV Ross still killed more than 60% of Vero-B4 4 days post infection (4dpi) in the MTS cell viability test. In a one-way ANOVA comparison of both CHIKV strains, no statistically significant difference could be detected with regard to cell infectivity and cell death between CHIKVBrazil and CHIKVRoss in Vero-B4 cells (Fig. 2A).
Effect of CHIKVRoss and CHIKVBrazil on different cell lines. Comparison of the infectivity/cell damage caused by two CHIKV strains CHIKVBrazil/Ross at increasing MOI. Cell viability was measured in a colorimetric assay (MTS cell viability test) 4dpi. Data are means ± SD of at least three independent experiments with three technical replicates, with 100% corresponding to non-infected cells (Mock). Asterisks indicating the p-values generated in a one-way ANOVA test comparison of non-infected cells with infected cells (green asterisks), and of the different virus strains at the same MOI (grey area and black asterisks). p-values are indicated as follows: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. A Vero-B4 cells (1 × 104 cells/well); B A549 cells (1 × 104 cells/well); C Huh-7 cells (5 × 103 cells/well); D DBTRG cells (1 × 104 cells/well); E U138 cells (1 × 104 cells/well); F U251 cells (1 × 104 cells/well)
A549 did not show any cytopathogenic effects (CPE) when infected with either CHIKV strain (Fig. 2B). Only at the highest MOI (6.45) with CHIKVRoss, limited cell death could be observed (65.96% ± 11.74% viable cells). Infection with a MOI of 63.5 and 6.35 of wt CHIKV even indicated proliferating cells (> 100% viable cells).
The human hepatoma cell line Huh-7 only showed cell death when infected with wt CHIKVBrazil at a very high MOI of 127 (Fig. 2C). Infection with MOI of 12.7 and lower did not result in a statistically different cell viability than non-infected Huh-7 cells. Infection with CHIKVRoss resulted in extensive cell death 4dpi when a MOI was between 0.0129 and 12.9 (80% dead Huh-7 cells). CHIKVRoss infection at a MOI 0.00129 still killed 45% of Huh-7 cells 4dpi. The comparison of the two CHIKV strains at corresponding MOI displayed a highly significant difference with p < 0.0001 between 0.0129 and 12.9 (Fig. 2C).
The brain derived cell line DBTRG was susceptible to CHIKV infection in a dose-dependent manner. At MOI ≥ 0.064, both virus strains showed diminished cell viability that was statistically significant (p < 0.0001) from non-infected cells (Fig. 2D). The wt CHIKVBrazil had similar significance at MOI 0.00064. High MOI (≥ 6.4) of both virus strains were needed to achieve extensive cell death > 50%.
The U138 cell line was susceptible to CHIKV infection and the cells showed extensive CPE 4dpi with either CHIKV strain. CHIKVRoss showed significantly more dead cells at a MOI 0.064 than CHIKVBrazil (35.5% vs. 56.2% surviving cells; p < 0.001). Likewise at a MOI of 0.64, 32% of the U138 cells survived CHIKVBrazil whilst 21% survived CHIKVRoss (Fig. 2E).
U138 did not show plaques when infected with CHIKVBrazil, although the plaque assays with U138 were conducted the same way as with Vero-B4.
U251 cells were more sensitive to CHIKV infection than U138 cells. At a MOI of 0.00064 of CHIKV Ross only 30.69 ± 18.46% of U251 cells survived after 4 days. There is however, no MOI dependent linear progression of the curve but rather an undulated one as far as CHIKVRoss on U251 is concerned (Fig. 2F). Four days after infection of U251 cells with CHIKVBrazil at MOI 0.00064, 56.32 ± 25.64% of the cells had survived. CHIKVBrazil at MOI ≥ 0.0064 kills > 65–70% of the U251 cells.
Comparison antiviral compounds vs virus/cell line
In Vero-B4 cells none of the administered compounds displayed any efficacy against either CHIKV strain (MOI: 0.64) at the applied concentrations (Fig. 3A). In U138 cells, RBV (410 µM), T-1105 (50 µM), and HCQ (10 µM) showed statistically significant efficacy against both CHIKV strains (p < 0.0001) (Fig. 3C). RBV and HCQ protected U138 cells significantly better from CPE caused by wt CHIKVBrazil than from the lab strain CHIKVRoss (p < 0.001 and < 0.0001, respectively).
Comparison of compound efficacy and toxicity against CHIKVRoss and CHIKVBrazil in the cell lines Vero-B4 and U138. Cells were treated with certain concentrations of HCQ, RBV, or T-1105 and were either infected with CHIKV (efficacy test A and C) or not (toxicity test B and D). Four days after infection/treatment, cell survival was determined with MTS. Values are given as percentages in relation to Mock control and are means of three independent experiments each with at least three technical replicates. A Vero-B4 and C U138 cells were infected with CHIKVRoss (white columns) or CHIKVBrazil (grey columns). Statistically significant differences of the compound efficacies between the different virus strains CHIKVRoss and wt CHIKVBrazil in the same cell line were evaluated in a one-way ANOVA test and are indicated by black asterisks. Red asterisks indicate significant (positive) differences between the positive control (black and grey line) and treated, infected cells (same corresponding virus strain and cell line). B and D Compound toxicity in Vero-B4 (B) and U138 (D) cells. Statistically significant (negative) differences between Mock control (grey bar and green line) and the treated cells (white bars), are indicated by blue asterisks. The number of asterisks indicate p-values as follows: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
In the toxicity testing Vero-B4 cells treated with T-1105, and HCQ showed a low toxic effect of the compound with 80–90% (± 5.31–9.45%) of the cells surviving (Fig. 3C). HCQ showed a highly significant difference to untreated cells with a cell survival of 81.44 ± 5.31% and p < 0.0001. RBV treatment resulted in more viable Vero-B4 cells than the untreated control (121.82 ± 15.57% viable cells), whilst in U138, RBV lead to statistically significant toxicity (70.43 ± 13.14% viable cells) (Fig. 3B, D). The difference in RBV toxicity between the two cell lines was statistically significant with p < 0.0001. Neither T-1105 nor HCQ led to significant cell damage in U138 cells (Fig. 3D).
IC 50 /CC 50 evaluation of RDV , HCQ, and T-1105 in Vero-B4 and U138 cells
A dose-dependent inhibition of CPE in both cell lines could be observed with HCQ and RBV. However, only in U138 cells was a dose-dependent effect for T-1105 detectable. Efficacies of HCQ and RBV differed considerably in the two cell lines (Table 1).
Of the four tested potential CHIKV antiviral substances (RBV, HCQ, and T-1105) only HCQ and RBV showed dose-dependent efficacies in Vero-B4 cells. However, even at 1000 µM concentration of RBV, only 37.55 ± 6.15% (at MOI 0.325) surviving cells were detectable and thus no IC50 value could be generated (data not shown). Efficacy of HCQ was observable between the concentrations of 1 µM and 30 µM. At concentrations > 30 µM HCQ was considerably toxic. An IC50 value of 18.29 µM and a CC50 of 49.63 µM could be generated for HCQ in Vero-B4 cells at MOI 0.355 (Fig. 4A and B) leading to an SI of 2.7. For T-1105 no dose-dependent efficacy against CHIKV in Vero-B4 could be observed (concentration range 5–100 µM).
IC50 and CC50 of HCQ in Vero-B4 and U138 cells. Hydroxychloroquine inhibits CHIKVBrazil-induced cell death in Vero-B4 (A) and U138 (C) cells in a dose-dependent manner. Cells (1 × 104 cells/well) were infected at an MOI of 0.355 and treated with a serial dilution of HCQ. After 4 days, cell death was determined via a colorimetric cell viability assay (MTS). Toxicity assays in Vero-B4 (B) and U138 (C) cells were performed similarly without infection of the cells. The data represent means ± SD of raw data from at least 3 independent experiments performed with three technical replicates. Normalised fit of dose–response curves was calculated with GraphPad Prism 6 Software
All tested compounds showed a dose-dependent antiviral effect against wt CHIKVBrazil in U138 cells. Efficacy of HCQ in U138 was observed using the compound at concentrations between 1 and 15 µM, as concentrations above 15 µM were toxic to the cells. An IC50 value of 4.136 µM and a CC50 of 35.45 µM was observed (Fig. 4C, D), leading to an SI of 8.57 for HCQ in U138.
RBV was effective against CHIKVBrazil in U138 cells with an IC50 of 165.8 µM (See Fig. 3A and B in Supplemental materials). No maximal toxic effect was observable at the highest concentration of 500 µM (data not shown). Consequently it was not possible to generate an exact CC50 value. As CC50 is > 500 µM, the SI would therefore be > 3.
The compound T-1105 (the defluorinated analogue of favipiravir) was effective against wt CHIKV in U138 cells with an IC50 of 34.21 µM (see supplemental materials). At the highest concentration (100 µM), no significant CPE was observable. The SI can thus be assumed to be > 3.
Taken together, these data demonstrate that HCQ, RBV, and T-1105 inhibit CHIKV-induced cell death of U138 cells in a dose-dependent manner. With the exception of HCQ the compounds had no significant toxic effect on this particular cell line at the tested concentrations.
Discussion
Kill curve experiments
CHIKV in vitro experiments are usually conducted in Vero cells as they propagate the virus well and show extensive CPE [23]. However, Vero cells originate from the kidney of an African green monkey and do not represent the usual site of infection in humans. As the latest CHIKV outbreaks reported an increase in neurological complications following CHIKF, it was one of our objectives to find a human derived neurological (immortalised) cell line to establish an in vitro model for neurogenic CHIKV (and possibly other neurogenetic alphavirus) infection.
There is a report of another glioblastoma cell line (U-87 MG (ATCC HTB-14)) being tested in CHIKV experiments [24, 25]. The study of Abraham et al. evaluated the glioblastoma cell line (U87-MG) with wt CHIKV isolate (RGCB355/KL08 CHIKV strain) with regard to susceptibility to infection, visible CPE, autophagy, apoptosis, and innate immune response. However, there are indications that this cell line is not the original cell line published by Ponten in 1968 [26, 27]. The DNA profile of the U87MG is different from that of the original and thus the origin of this cell line is unknown [25].
For these reasons, we tested different human glioblastoma cell lines (DBTRG, U138, and U251) for the susceptibility of infection with CHIKV and their suitability for cell viability assays with this virus. Furthermore, Huh-7 and A549, for which controversial data with regard to CHIKV infectivity have been published, were evaluated with the same objectives. As these differences might be due to the fact that different CHIKV strains have been used in the aforementioned studies, we compared the lab-adapted CHIKV strain Ross and the field isolate from Brazil in in vitro cell cultures and by full genome analysis.
In our study, all tested glioblastoma cell lines were susceptible to CHIKV infection. However, in DBTRG cells, extensive CPE with > 50% nonviable cells could only be achieved at MOI ≥ 6.4. U138 and U251 cells showed extensive CPE 4dpi with either CHIKV strain (Fig. 2E). U251 cells were more sensitive to wt CHIKVBrazil infection than U138 cells. Yet, the kill curve of U251 infected with CHIKVRoss strain was not a strictly dose-dependent linear progression but rather an undulated one (Fig. 2F). Furthermore, SD in U251 was also rather high (sometimes > 25%).
One important observation was that cell viability assay with MTS in U251 is not working reliably when the experiment duration exceeds 3 days and the initial cell concentration is ≥ 1 × 104 cells/well. The reason might be that the cells double in 23 h and are sensitive to overgrowing [26, 28]. Too many cells will cause U251 to stop proliferating, curb their metabolic rates, and reach a state of stasis. In this state, U251 cells no longer reduce MTS into its formazan product. Consequently, the absorbance of the plate appears to be the same as in dead cells although there are a multitude of alive U251 cells. This results in false interpretation of test results. Seeding too few cells, on the other hand, results in badly proliferating cells, since it was our observation that both U251 (and U138) cells need close cell-to-cell contacts in order to form a stable layer. For these reasons, U251 were not used in the other experiments, since those experiments were designed to run for 4 days. Still, U251 cells might be a useful cell line for CHIKV studies if the experimental parameters are adapted accordingly.
The human lung derived cell line A549 proved unsuitable experiments testing cell viability since it displayed no CPE after infection with wt CHIKVBrazil and only limited cell death at the highest MOI (6.45) of CHIKVRoss at 4dpi (Fig. 2B). The kill curve in MTS assays of wt CHIKVBrazil on A549 cells even indicated more viable cells at the highest MOI (63.5 and 6.35) than in the non-infected control. This phenomenon might be explained with some of the cells dying at such high MOI (possibly due to apoptosis), which then leaves more space and substrate for the surviving cells. At lower MOI and in the control, the A549 cells were overconfluent and might have curbed down their metabolism, leading to a lower MTS reduction (which leads to lower OD values). Apart from a bad or unreliable CPE, the amount of virus needed to conduct viability experiments on A549 is very high. The A549 cell line has been described in CHIKV experiments before, but reports are contradictory. Sourisseau, Schilte [29] state that wt CHIKV virions bind to A549 cells without replicating within the cell, and Solignat, Gay [30] did not observe any CPE on wt CHIKV infected A549. Other studies do not recommend this cell line claiming that CHIKV does not reproduce in A549 [31]. Franco, Rodriquez [32], however, used this cell line to test RBV and favipiravir against an attenuated CHIKV strain (vaccine strain 181/clone25) at MOI 0.1 in a yield assay, looking at virus in the supernatant. This would indicate that this particular CHIKV strain does replicate in A549 cells and is secreted into the supernatant. It is possible that the laboratory-generated, attenuated vaccine strain (181/clone25) has some affinity to this cell line, yet, for cell viability experiments with our clinical isolate of CHIKVBrazil and the Ross strain, A549 cell cannot be recommended.
The Huh-7 human hepatocarcinoma cell line is often used to evaluate hepatocellular toxicity of compounds in vitro [33]. Huh-7 cells only showed cell death after infection with wt CHIKVBrazil at a very high MOI of 127. Data showed that an increased initial MOI of CHIKV promotes the effect of CHIKV-induced cellular transcriptional shutoff in cells and thus leads to apoptosis [34]. This effect could be observed in cells infected with higher MOI [34], and it could explain the CPE in A549 and Huh-7 at very high MOI. We observed the biggest difference in CPE between the two virus strains in Huh-7 cells. Whilst the wt CHIKV was not able to significantly damage Huh-7 cells at MOI ≤ 12.7, the Ross strain showed a dose-dependent CPE (Fig. 2C).
Solignat, Gay [30] has successfully used Huh-7 cells in CHIKV experiments before. In his work, Huh-7 cells were infected at higher MOI with the West African CHIKV strain 5′CHIKV-EGFP that encodes a GFP protein. According to the study, there was detectable viral replication and CPE [30]. Antiviral efficacy assays measuring virus yield were conducted using Huh-7 cells by Franco, Rodriquez [32] (vaccine strain of CHIKV (181/clone 25)) and Ferreira, Reis [35] (CHIKV (Asian strain), not further specified). Additionally, a study from Roberts, Zothner [36] evaluated a variety of cell lines for their use in experiments with a sub-genomic replicon (SGR) system CHIKV SGR (CHIKV-D-Luc-SGR), derived from the ECSA strain (ICRES). To test infectious virus, the group used a full-length infectious cDNA clone of CHIKV-LR2006 OPY1. According to the group, Huh-7 cells could be infected by said CHIKV construct and did yield infectious virus in moderate amounts. A549 cells on the other hand were less suited. No observations were done in regard of CPE in the two cell lines in this particular study. Thus, the results of the research cannot be transferred to cell viability assays with wt CHIKV.
The fact that other studies have successfully used the Huh-7 cell line in CHIKV cell viability assays might be due to the use of different, lab-adapted, or modified CHIKV strains. Interestingly, the field isolate tested in this study showed no CPE on Huh-7 cells whilst the Ross strain displayed extended cell kill. This might be due to cell culture adaption of CHIKVRoss. Genome analysis of both strains revealed that both CHIKV strains (CHIKVRoss and CHIKVBrazil) belong to the ESCA clade.
For other arboviruses like Dengue Virus (DENV) or Zika Virus (ZIKV), A549, and Huh-7 are very useful cell lines, as these viruses replicate well and show CPE [31, 37,38,39]. Since coinfections of CHIKV, DENV, and ZIKV occur due to geographical overlapping in tropical regions, cell lines in which all these viruses may be propagated might have been one objective as to why A549 and Huh-7 cells have repeatedly been tried in CHIKV experiments. Especially DENV and CHIKV cause similar fever-like symptoms, and are difficult to diagnose [31].
To our knowledge a comparison of lab-adapted CHIKV strain with wt CHIKV isolates with regard to cell affinity in different cell lines has only been done by Wikan, Sakoonwatanyoo [40]. The group tested a panel of cell lines with different CHIKV strains (two field isolates and the original Ross strain). Still, their cell line panel did not encompass Huh-7 and A549 cells.
The reasons for the different CPE of CHIKVBrazil and CHIKVRoss on various cell lines are currently unknown. One possible explanation might be the presence or absence of specific cell surface receptors and/or host proteins which are necessary for an efficient infection, replication, and virus production with cell lysis. Even if certain cell lines have already been described as susceptible, different CHIKV strains might still not work.
Various studies observed strain differences in CHIKV tropism and virulence [41]. The cell culture adapted CHIKV strain 181/25, which had been investigated as a possible vaccine strain after being passaged various times in vitro, displays increased glycosaminoglycan (GAG) binding due to a specific mutation in the E2 glycoprotein (G82R) [41,42,43,44,45]. GAGs serve as attachment factors for many pathogenic viruses and are amongst the central factors which trigger CHIKV attachment [44]. The viral spike glycoproteins E2 and E1 play an important role for the infection of target cells. Whilst the E2 protein is thought to be responsible for receptor binding, the E1 protein contains a hydrophobic fusion peptide and is necessary for viral and cellular membrane fusion [46].
da Silva and colleagues could demonstrate by reciprocal amino acid substitutions at residue 82 of the E2 glycoprotein that the exchange G82R resulted in a phenotype switch in CHIKV [44]. Their data suggest that an Arginine at position 82 of E2 increases the affinity of the glycoprotein for GAGs [44]. These findings also support the hypothesis that the G82R substitution in E2 of CHIKV strain 181/25 contributes to attenuation of the vaccine strain due to GAG binding [45]. Further research in vitro and in vivo conclude that an arginine at residue 82 lead to a greater dependence on GAGs for infection of mammalian cells [41]. These results indicate that GAG utilisation plays a role in regulating CHIKV tropism and host responses that contribute to arthritis, a cardinal symptom of CHIKV disease [41].
Other point mutations in the E2 protein (e.g. E79K, E266K, and E166K) affecting GAG binding were observed in cell culture adapted CHIKV strains [43, 47, 48]. These strains were more dependent on GAGs for infection and showed reduced in vivo replication. By increasing the positive charge in domain A of the E2 protein, these point mutations affected the binding affinity of the virus. The positive charge acquisition is a phenomenon commonly observed in cell culture adapted alphaviruses and often correlates with an attenuated phenotype in vivo [45, 49, 50].
Mutation at critical points of the envelope surface proteins may introduce changes in charge and hydrophobicity of the CHIKV E1 and E2 glycoprotein [51]. Such changes in the E1/E2 proteins can influence pH sensitivity and dramatically affect virus structure and production [52, 53]. Furthermore, mutations in specific regions of the E2 protein may directly influence interactions with a specific cell surface receptor thus influencing virulence and adaption [48, 54].
Whole genome sequencing revealed 5 differences in the E1 glycoprotein (Fig. 1). One difference was at the E1 protein position 322. Whilst CHIKVRoss has a valine in this position, CHIKVBrazil has an alanine. Studies showed that membrane fusion of endosomes containing CHIKV is triggered by E1 glycoproteins and that this process is pH dependent. Mutations in the E1 protein at position 226 can lead to phenotypes which require lower pH compared to the parent strains to trigger fusion [55, 56].
Differences in the E1 protein between the two strains may be responsible for the differences in HCQ response, as HCQ (and the more toxic base substance chloroquine (CQ)) is known to raise the endosomal pH and thus intervene with CHIKV membrane fusion [57]. It is therefore possible that some of these changes have an impact on the acid pH-triggered conformational changes in alphavirus E1 during membrane fusion [58].
Furthermore, whole genome sequencing of the strains used in this study revealed differences at four positions in the nsP2, a protein known to be connected with cytopathogenicity especially of old-world alphaviruses. Apart from other functions, the nsP2 inhibits host transcription which eventually induces cell death [59].
Whether the discovered genome differences between CHIKVRoss and CHIKVBrazil are responsible for the differences in cell affinity (especially concerning Huh-7 cells) need to be further investigated using mutagenesis of the respective sites and observation on the effects on cell tropism in reverse genetics experiments.
Comparison of compound efficacy in different cell lines against two different CHIKV strains
Despite being treated with compounds that should potentially confer some protection at the concentrations used, Vero-B4 cells showed no significant cell survival after 4 days of CHIKV challenge. A possible reason for the inefficacy of the compounds might be the higher MOI of 0.64 with which the cells were infected (compared to an MOI of 0.355 in the IC50/CC50 experiments and considerably lower MOIs of 0.005–0.01 in previous studies with the same setup [21].
Previously published data states that RBV was efficient against wt CHIKV (MOI: 0.005) with an IC50 of 423.6 µM and a CC50 > 500 µM [21]. The same study states CQ’s IC50 as 5–10.6 µM with a CC50 of > 36 µM. Delang, Segura Guerrero [22] however tested CQ against CHIKV Indian Ocean strain 899 (lab) at MOI 0.01 in Vero cells and generated IC50 values of 11 and 28 µM. Delang also tested T-1105 against this lab CHIKV strain at MOI 0.01 and IC50 values were 7–47 µM, with a CC50 value of 571 µM [22]. HCQ is a less toxic derivative of CQ and its efficacy is comparable to CQ.
U138 cells on the other hand benefited considerably from RBV, T-1105 (50 µM), and HCQ treatment, despite the higher MOI. The reason for the difference in compound efficacy between the two cell lines might be due to the different ability of the respective cells to process the compounds into their active analogues.
Furthermore, there was a significant difference in RBV toxicity between the two cell lines. Whilst RBV lead to an increase of the MTS signal in Vero-B4 cells, U138 cells showed diminished signals which can be interpreted as fewer viable cells. There might be different reasons for this observation:
-
i.
The compounds kill some cells, leave space for the remaining cells which then have spare room and medium and become highly metabolically active, hence they are able to reduce MTS into the signal yielding formazan product more effectively.
-
ii.
Vero-B4 have a higher proliferation rate (doubling time 24 h) than U138 cells (doubling time 47–72 h) [28, 60]. It is thus possible that Vero-B4 cells also have a higher metabolism and are able to process RBV quicker into a less toxic compound.
-
iii.
Additionally, there is the chance that RBV actually causes cell proliferation or an activation of metabolism in Vero-B4 cells, whilst U138 cells are hampered/damaged by the compound.
When comparing efficacies of the compounds between the two strains, RBV and HCQ protected U138 cells significantly better from wt CHIKV than from CHIKV Ross.
CQ/HCQ are effective at early stages of viral infection [61]. The drugs seem to impair cell-virus surface interactions. Pre-treatment of Vero cells with CQ impairs terminal glycosylation of ACE2, a cell surface receptor used by severe acute respiratory syndrome corona virus (SARS-CoV) for cell attachment [62]. Khan et al. suggested a similar mechanism to be responsible for the inhibition of CHIKV infection by CQ in vitro [61]. In the case of other alphaviruses like Sindbis virus (SINV) and Semliki Forest virus (SFV), viral fusion with the host cell membrane is achieved via conformational changes in the viral envelope glycoprotein. These changes are triggered by clathrin-mediated endocytosis by the target cell and the low pH of the endosomal compartment [63]. This low endosomal pH is said to be required for CHIKV entry into cells as well [29]. Bernard and colleagues could demonstrate that the base CQ raises the endosomal pH by interfering with the protonation of the endocytic vesicles. This prevents the E1 fusion step needed for the release of CHIKV RNA into the cell cytoplasm [64].
In our comparative experiments, HCQ showed a statistically significant higher efficacy against the wt CHIKVBrazil than against the CHIKVRoss strain. The CHIKVBrazil strain may rely on a lower pH to grant membrane fusion (possibly due to mutations in the E1 glycoprotein as mentioned above), or the strain CHIKVRoss has gained a more efficient way to grant fusion with the host cell membrane during its repeated passage in Vero cells (possibly due to mutations in the E2 protein). It should be mentioned that HCQ is only used as a control for measuring efficacy in vitro, as patients do not benefit from HCQ treatment during acute CHIKV disease and the drug has no suppressive effect on peripheral viral load in patients [65].
Differences of IC50/CC50 values in different cell lines
Both, IC50 and CC50 of HCQ observed in this study are higher than previously published data of chloroquine in Vero cells. This might be due to a different MOI.
RBV did show a dose-dependent efficacy, however, the maximal protection of Vero-B4 cells at the highest drug concentrations did not outnumber 37.55 ± 6.15% (at MOI 0.325) surviving cells and thus no IC50 value could be generated. Published data from comparable experiments give IC50 values for RBV of 423.6–765.8 µM in Vero-E6 cells [6, 21]. One possible explanation for not exceeding 37.55% surviving Vero-B4 cells might be the fact that the aforementioned publication used different CHIKV strains, VeroE6 cells, and infected with a lower MOI (0.005). At the highest concentration (1000 µM) RBV showed no toxic effect on Vero-B4 cells. The other compounds neither displayed a positive effect against CHIKVBrazil nor negative effects on Vero-B4 cells at the used concentrations. Altogether, the experiments showed that HCQ and RBV inhibit CHIKVBrazil-induced cell death of Vero-B4 cells in a dose-dependent manner and that HCQ was considerably more effective in preventing CHIKV-related CPE in Vero-B4 than RBV (Table 1).
Vero-B4 cells could not be protected from CHIKV infection with T-1105 at the concentrations used. This was unexpected, since Delang reported IC50 values of 7–47 µM for T-1105 in Vero cells in his study [22]. The concentrations used in the experiments for T-1105 in this study ranged from 5 to 100 µM, well in the range to detect an efficacy of the compound against CHIKVBrazil. However, Delang used VeroA cells, different CHIKV strains and infected the cells with an MOI of 0.1. It is possible that the difference in CHIKV strain, cell line, and MOI contributed to the discrepancy between our results and previously published data. Since the compound did show efficacy against CHIKVBrazil in U138 cells, issues related to the compound itself (e.g. degradation due to repeated thaw-freeze-cycles) can be ruled out.
Both RBV and T-1105 are antivirals that interfere with the viral genome replication by inhibiting the nsP4 polymerase. Both are synthetic purine nucleoside analogues [6], and act as broad-spectrum antivirals, with multiple mechanisms of action ascribed to them. Both might either block the RNA-dependent RNA polymerase (RdRp) function of the nsP4 by binding at certain domains of the enzyme and/or they might be incorporated into the viral genome and thus lead to lethal mutagenesis [32]. Others suggest that RBV interferes with the nsP1 guanylyl transferase and/or methyltransferase activity and thus leads to a production of untranslatable mRNAs [66]. RBV and T-1105 (as well as the fluorinated form favipiravir T-705) have to be phosphorylated by host cell kinases into their mono-, di-, and triphosphate metabolites. The triphosphate form is the active metabolite which is eventually incorporated into the viral genome, thus leading to error catastrophe [67].
Resistance against RBV and favipiravir (T-705) has been reported and is explained by mutations in nsP4. RBV resistance was put down to a mutation from K291R in nsP4 whilst favipiravir resistance was explained by a C483Y mutation [22, 66]. Whole genome sequencing of our strains revealed that neither CHIKV Ross nor Brazil have these mutations. Our experiments confirmed the findings of Franco and colleagues that compound efficacy varies between host cell lines. While Vero-B4 cells were refractory to the treatment of RDV, T-1105, and to a lesser extend HCQ, U138 cells could be protected by all three compounds considerably better. A study demonstrated that the accumulation of RBV is host cell dependent due to the presence or absence of specific nucleoside transporters [68]. This could also hold true for other nucleoside analogues like T-1105. Furthermore, pro-drugs like RBV and T-1105 depend on host kinases for phosphorylation into their active metabolite. The resistance of some cell types to RBV may thus depend on the intracellular RBV metabolism [69]. A study on the cell line-dependent activation and antiviral activity of T-1105 revealed that T-1105 activation in Vero cells was hindered by inefficient conversion of the ribonucleoside monophosphate to the ribonucleoside diphosphate en route to forming the active triphosphate [70]. This might be one reason, why T-1105 is less potent in Vero-B4 than in U138 cells. It is likely that the distribution of host cell kinases differs between species and tissues and thus lead to a varying intracellular concentration of the triphosphate forms of RBV and possibly T-1105 [32].
Conclusion
Two glioblastoma cell lines (U138 and U251) were identified as potentially useful in vitro cell culture models for CHIKV infection and evaluation of antiviral activity. To our knowledge, this is the first time these two cell lines have been described in connection with CHIKV antiviral tests. Furthermore, A549 and Huh-7 cells cannot be recommended for cell viability assays with wt CHIKV, as these cell lines do not show CPE. Furthermore, our experiments proved that there are differences in cytopathological effects and antiviral efficacies between wt and laboratory-adapted CHIKV strains.
Abbreviations
- 95% CI:
-
95% Confidence interval
- Abs:
-
Absorption
- CC50 :
-
Half maximal cytotoxic concentration
- CHIKF:
-
Chikungunya fever
- CHIKV:
-
Chikungunya virus
- Cp:
-
Capsid protein
- CPE:
-
Cytopathic effect
- CQ:
-
Chloroquine
- DENV:
-
Dengue virus
- dpi:
-
Days post infection
- ECSA:
-
East-Central-South African
- FBS:
-
Foetal bovine serum
- FDA:
-
U.S. Food and Drug Administration
- GAG:
-
Glycosaminoglycans
- HCQ:
-
Hydroxychloroquine
- HG:
-
“High glucose”; medium supplemented with 4.5 g/L of d-glucose
- IC50 :
-
Half maximal inhibitory concentration
- IOL:
-
Indian Ocean Lineage
- LG:
-
“Low glucose”; medium supplemented with 1 g/L of d-glucose
- MOI:
-
Multiplicity of infection
- n:
-
Number of independent repetitions
- NC:
-
Nucleocapsid
- nsp:
-
Non-structural protein
- RBV:
-
Ribavirin
- RdRp:
-
RNA-dependent RNA polymerase
- SD:
-
Standard deviation
- SFV:
-
Semliki Forest virus
- SI:
-
Selectivity index
- SINV:
-
Sindbis virus
- WA:
-
West African
- wt:
-
Wild type
- ZIKV:
-
Zika virus
References
Singh SK (2015) Overview on chikungunya virus pathogenesis. In: Singh SK (ed) Human emerging and re-emerging infections: viral & parasitic infections. Wiley, New Jersey, pp 177–188
Powers AM et al (2000) Re-emergence of Chikungunya and O’nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol 81(Pt 2):471–479
Schuffenecker I et al (2006) Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med 3(7):e263
Brown RS, Wan JJ, Kielian M (2018) The alphavirus exit pathway: what we know and what we wish we knew. Viruses 10(2):89
Hucke FIL, Bestehorn-Willmann M, Bugert JJ (2020) Prophylactic strategies to control Chikungunya virus infection. Virus Genes 57:133
Hucke FIL, Bugert JJ (2020) Current and promising antivirals against chikungunya virus. Front Public Health 8:916
Matusali G et al (2019) Tropism of the Chikungunya virus. Viruses 11(2):175
Zhang R et al (2018) Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557(7706):570–574
Ruiz Silva M et al (2016) Mechanism and role of MCP-1 upregulation upon chikungunya virus infection in human peripheral blood mononuclear cells. Sci Rep 6:32288
Lokireddy S, Vemula S, Vadde R (2008) Connective tissue metabolism in chikungunya patients. Virol J 5:31
Mehta R et al (2018) The neurological complications of chikungunya virus: a systematic review. Rev Med Virol 28(3):e1978
Cerny T et al (2017) The range of neurological complications in Chikungunya fever. Neurocrit Care 27(3):447–457
Das T et al (2015) Multifaceted innate immune responses engaged by astrocytes, microglia and resident dendritic cells against Chikungunya neuroinfection. J Gen Virol 96(Pt 2):294–310
Yasumura YK (1963) Studies on SV40 in tissue culture-preliminary step for cancer research in vitro. Nihon Rinsho 21:1201–1215
Lieber M et al (1976) A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer 17(1):62–70
Nakabayashi H et al (1982) Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res 42(9):3858–3863
Kruse CA et al (1992) Characterization of a continuous human glioma cell line DBTRG-05MG: growth kinetics, karyotype, receptor expression, and tumor suppressor gene analyses. In Vitro Cell Dev Biol 28(9–10):609–614
Ross RW (1956) The Newala epidemic. III. The virus: isolation, pathogenic properties and relationship to the epidemic. J Hyg (Lond) 54(2):177–191
Arankalle VA et al (2007) Genetic divergence of Chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. J Gen Virol 88(Pt 7):1967–1976
Volk SM et al (2010) Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J Virol 84(13):6497–6504
Scholte FE et al (2013) Characterization of synthetic Chikungunya viruses based on the consensus sequence of recent E1–226V isolates. PLoS ONE 8(8):e71047
Delang L et al (2014) Mutations in the chikungunya virus non-structural proteins cause resistance to favipiravir (T-705), a broad-spectrum antiviral. J Antimicrob Chemother 69(10):2770–2784
Sudeep AB et al (2019) Differential susceptibility & replication potential of Vero E6, BHK-21, RD, A-549, C6/36 cells & Aedes aegypti mosquitoes to three strains of chikungunya virus. Indian J Med Res 149(6):771–777
Abraham R et al (2013) Induction of cytopathogenicity in human glioblastoma cells by chikungunya virus. PLoS ONE 8(9):e75854
Allen M et al (2016) Origin of the U87MG glioma cell line: good news and bad news. Sci Transl Med 8(354):35re43
Pontén J, Macintyre EH (1968) Long term culture of normal and neoplastic human glia. Acta Pathol Microbiol Scand 74(4):465–486
Pontén J, Westermark B (1978) Properties of human malignant glioma cells in vitro. Med Biol 56(4):184–193
Weller M et al (1998) Predicting chemoresistance in human malignant glioma cells: the role of molecular genetic analyses. Int J Cancer 79(6):640–644
Sourisseau M et al (2007) Characterization of reemerging chikungunya virus. PLoS Pathog 3(6):e89
Solignat M et al (2009) Replication cycle of chikungunya: a re-emerging arbovirus. Virology 393(2):183–197
Olagnier D et al (2014) Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type I interferon-independent stimulation of the innate antiviral response. J Virol 88(8):4180–4194
Franco EJ et al (2018) The effectiveness of antiviral agents with broad-spectrum activity against chikungunya virus varies between host cell lines. Antivir Chem Chemother 26:2040206618807580
Lin J et al (2012) Comparative analysis of phase I and II enzyme activities in 5 hepatic cell lines identifies Huh-7 and HCC-T cells with the highest potential to study drug metabolism. Arch Toxicol 86(1):87–95
Li YG et al (2013) Chikungunya virus induces a more moderate cytopathic effect in mosquito cells than in mammalian cells. Intervirology 56(1):6–12
Ferreira AC et al (2019) Beyond members of the flaviviridae family, sofosbuvir also inhibits chikungunya virus replication. Antimicrob Agents Chemother 63(2):e01389
Roberts GC et al (2017) Evaluation of a range of mammalian and mosquito cell lines for use in Chikungunya virus research. Sci Rep 7(1):14641–14641
Chan JF et al (2016) Differential cell line susceptibility to the emerging Zika virus: implications for disease pathogenesis, non-vector-borne human transmission and animal reservoirs. Emerg Microbes Infect 5(8):e93
Vicenti I et al (2018) Comparative analysis of different cell systems for Zika virus (ZIKV) propagation and evaluation of anti-ZIKV compounds in vitro. Virus Res 244:64–70
Franco EJ, Pires de Mello CP, Brown AN (2021) Antiviral evaluation of UV-4B and interferon-alpha combination regimens against dengue virus. Viruses 13(5):771
Wikan N et al (2012) Chikungunya virus infection of cell lines: analysis of the East, Central and South African lineage. PLoS ONE 7(1):e31102
Ashbrook AW et al (2014) Residue 82 of the Chikungunya virus E2 attachment protein modulates viral dissemination and arthritis in mice. J Virol 88(21):12180–12192
Levitt NH et al (1986) Development of an attenuated strain of chikungunya virus for use in vaccine production. Vaccine 4(3):157–162
Gardner CL et al (2014) Deliberate attenuation of chikungunya virus by adaptation to heparan sulfate-dependent infectivity: a model for rational arboviral vaccine design. PLoS Negl Trop Dis 8(2):e2719
Silva LA et al (2014) A single-amino-acid polymorphism in Chikungunya virus E2 glycoprotein influences glycosaminoglycan utilization. J Virol 88(5):2385–2397
Gorchakov R et al (2012) Attenuation of Chikungunya virus vaccine strain 181/clone 25 is determined by two amino acid substitutions in the E2 envelope glycoprotein. J Virol 86(11):6084–6096
Schnierle BS (2019) Cellular attachment and entry factors for chikungunya virus. Viruses 11(11):1078
Henrik Gad H et al (2012) The E2–E166K substitution restores Chikungunya virus growth in OAS3 expressing cells by acting on viral entry. Virology 434(1):27–37
Coffey LL et al (2011) Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc Natl Acad Sci USA 108(38):16038–16043
Davis NL et al (1991) Attenuating mutations in the E2 glycoprotein gene of Venezuelan equine encephalitis virus: construction of single and multiple mutants in a full-length cDNA clone. Virology 183(1):20–31
Klimstra WB, Ryman KD, Johnston RE (1998) Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol 72(9):7357–7366
Maljkovic Berry I et al (2019) Global outbreaks and origins of a chikungunya virus variant carrying mutations which may increase fitness for Aedes aegypti: revelations from the 2016 Mandera, Kenya Outbreak. Am J Trop Med Hyg 100(5):1249–1257
Akahata W, Nabel GJ (2012) A specific domain of the Chikungunya virus E2 protein regulates particle formation in human cells: implications for alphavirus vaccine design. J Virol 86(16):8879–8883
Lu YE et al (2001) In vivo generation and characterization of a soluble form of the Semliki forest virus fusion protein. J Virol 75(17):8329–8339
Tsetsarkin KA, Weaver SC (2011) Sequential adaptive mutations enhance efficient vector switching by Chikungunya virus and its epidemic emergence. PLoS Pathog 7(12):e1002412–e1002412
Tsetsarkin KA, McGee CE, Higgs S (2011) Chikungunya virus adaptation to Aedes albopictus mosquitoes does not correlate with acquisition of cholesterol dependence or decreased pH threshold for fusion reaction. Virol J 8:376
Gay B et al (2012) pH-dependent entry of chikungunya virus into Aedes albopictus cells. Infect Genet Evol 12(6):1275–1281
Askarian F et al (2021) A review on the pharmacokinetic properties and toxicity considerations for chloroquine and hydroxychloroquine to potentially treat coronavirus patients. Toxicol Res. https://doi.org/10.1007/s43188-021-00101-5
Sahoo B, Gudigamolla NK, Chowdary TK (2020) Acidic pH-induced conformational changes in chikungunya virus fusion protein E1: a spring-twisted region in the domain I-III linker acts as a hinge point for swiveling motion of domains. J Virol 94(23):e01561-e1620
Akhrymuk I et al (2019) Novel mutations in nsP2 abolish chikungunya virus-induced transcriptional shutoff and make the virus less cytopathic without affecting its replication rates. J Virol 93(4):e02062-e2118
Ammerman NC, Beier-Sexton M, Azad AF (2008) Growth and maintenance of Vero cell lines. Curr Protoc Microbiol 4:4E
Khan M et al (2010) Assessment of in vitro prophylactic and therapeutic efficacy of chloroquine against Chikungunya virus in vero cells. J Med Virol 82(5):817–824
Vincent MJ et al (2005) Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J 2:69
DeTulleo L, Kirchhausen T (1998) The clathrin endocytic pathway in viral infection. Embo J 17(16):4585–4593
Bernard E et al (2010) Endocytosis of chikungunya virus into mammalian cells: role of clathrin and early endosomal compartments. PLoS ONE 5(7):e11479
Roques P et al (2018) Paradoxical effect of chloroquine treatment in enhancing chikungunya virus infection. Viruses 10(5):268
Beaucourt S, Vignuzzi M (2014) Ribavirin: a drug active against many viruses with multiple effects on virus replication and propagation. Molecular basis of ribavirin resistance. Curr Opin Virol 8:10–15
Crotty S, Cameron CE, Andino R (2001) RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci USA 98(12):6895–6900
Ibarra KD, Pfeiffer JK (2009) Reduced ribavirin antiviral efficacy via nucleoside transporter-mediated drug resistance. J Virol 83(9):4538–4547
Shah NR, Sunderland A, Grdzelishvili VZ (2010) Cell type mediated resistance of vesicular stomatitis virus and Sendai virus to ribavirin. PLoS ONE 5(6):e11265
Huchting J et al (2019) Cell line-dependent activation and antiviral activity of T-1105, the non-fluorinated analogue of T-705 (favipiravir). Antiviral Res 167:1–5
Acknowledgements
This work was funded by the Bundeswehr Medical Service’s Biodefence research programme and the DZIF TI 07.003 MD Programme-Hucke of the German Center for Infection Research (DZIF), as well as the Zoonoses Sequencing Network BMBF-ZooSeq (01KI1905A). Special thanks go to Dr. Ruth Brack-Werner of the Helmhotz Institute in Munich for a loan of the human glioblastoma cell lines U138 and U251.
Author information
Authors and Affiliations
Contributions
JJB and FH conceived the layout of the project. FH performed all experiments with CHIKV. MB-W was responsible for all work connected with sequencing and genome analysis. FH, MB-W and PZ performed statistical analysis and generated the figures and tables. FH and MB-W wrote the first draft of the manuscript. MB-W, MB, AB, and JJB contributed providing additional information as well as reviewing the manuscript. JJB supervised and funded the project as well as oversaw data analysis, manuscript drafting, and revision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest. The authors declare that there is no financial or personal relationship with other people or organisations that could inappropriately influence the work. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by Bundeswehr Joint Medical Service or any other governmental institutions.
Ethical approval
All human cell lines used in this study are commercially available and their origin have been stated in the Materials and Methods section. A wt CHIKV was isolated from a diagnostic sample. No patient metadata was collected. No studies with human participants were conducted. Patient consent was obtained according to the national rules for the collection and prior to performing the diagnostic investigation.
Additional information
Edited by A. Lorena Passarelli.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hucke, F.I.L., Bestehorn-Willmann, M., Bassetto, M. et al. CHIKV strains Brazil (wt) and Ross (lab-adapted) differ with regard to cell host range and antiviral sensitivity and show CPE in human glioblastoma cell lines U138 and U251. Virus Genes 58, 188–202 (2022). https://doi.org/10.1007/s11262-022-01892-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-022-01892-x