Abstract
The human parainfluenza virus type 3 (HPIV3) phosphoprotein (P) gene is unusual as it contains an editing site where nontemplated ribonucleotide residues can be inserted. This RNA editing can lead to the expression of the viral P, PD, putative W, and theoretical V protein from a single gene. Although the HPIV3 PD protein has been detected, its function and those of the W and V proteins are poorly understood. Therefore, we first used reverse genetics techniques to construct and rescue a recombinant (r)HPIV3 clone with a polyhistidine sequence at the 5′ end of the P gene for tagged protein detection. Western blot analysis demonstrated the presence of the P, PD, and W proteins, but no V protein was detected. Then, we functionally studied the D domain of the PD protein by constructing two rHPIV3 knockout clones that are deficient in the expression of the D domain. Results from growth kinetic studies with infected MA-104 and A596 cells showed that viral replication of the two knockout viruses (rHPIV3-ΔES and rHPIV3-ΔD) was comparable to that of the parental virus in both cell lines. However, viral mRNA transcription and genomic replication was significantly reduced. Furthermore, cytokine/chemokine profiles of A549 cells infected with either knockout virus were unchanged or showed lower levels compared to those from cells infected with the parental virus. These data suggest that the D domain of the PD protein may play a luxury role in HPIV3 RNA synthesis and may also be involved in disrupting the expression of beta interferon.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most members of the Paramyxovirinae subfamily, classified in the Paramyxoviridae family, express additional accessory proteins from the phosphoprotein (P) gene by mRNA editing. The mRNA editing phenomenon occurs during viral mRNA transcription when the viral RNA polymerase stops, stutters, and inserts nontemplated guanosine (G) residues into the mRNA transcript at an editing site located in the middle of the P gene [1, 2]. The result is a pool of P gene mRNAs with differing G insertions leading to the translation of fusion proteins with identical N-terminal ends and different C-terminal ends. Most paramyxoviruses express the P, V, and W proteins using this mRNA editing mechanism with the exception of human parainfluenza virus type 1 (HPIV1), which does not encode an editing site and, therefore, does not express the V or W accessory proteins [1–3]. The human parainfluenza virus type 3 (HPIV3), also a paramyxovirus, was shown to express yet another protein, the PD protein—sometimes referred to only as D protein, from the P gene using the same mRNA editing mechanism [4].
Although the HPIV3 PD protein has been detected, its function and those of the putative W and/or theoretical V viral proteins remain undefined. Wells and Malur [4] demonstrated that the PD protein was primarily localized to the nucleus of the host cells. They also identified three nuclear localization signals on the PD protein. Two of the three signals were found on the C-terminal domain of the PD protein, while the third signal was shared by all proteins expressed from the P gene. Durbin et al. [5] examined possible functions of the HPIV3 D and V domains by reverse genetics. They constructed HPIV3 knockout viruses in which the D and V domains were either silenced individually or simultaneously. No attenuation of viral titers was seen when the individual or simultaneous knockout viruses were replicated in vitro, individually in vivo, or simultaneously in nasal turbinates. However, attenuation was seen when the domains were silenced simultaneously and the knockout virus replicated in the lung. These data suggest that the V domain may be expressed by an unknown, tissue-specific mechanism and that the D and V domains may work in concert to counteract the antiviral response of the host cell.
The distinguishing feature of the paramyxovirus V protein is a conserved C-terminal cysteine-rich domain that binds two zinc atoms and forms a zinc-finger structure [6]. This feature is also encoded for in the HPIV3 P gene in the same reading frame as the W protein downstream of two or three stop codons, depending upon the virus strain [1, 7]. The main function of the paramyxovirus V protein is to inhibit the primary antiviral response by binding to either melanoma differentiation-associated gene 5 (MDA5), thus preventing viral dsRNA binding, or alpha I-kappa B kinase (IKKα), thereby preventing phosphorylation of interferon regulatory factor 7 (IRF7) [8–10]. In either case, the V protein ultimately interferes with the primary antiviral signaling pathway and inhibits the expression of beta interferon (IFN-β).
This study served two purposes. Although the HPIV3 genome may contain the nucleotide sequence that encodes for a cysteine-rich V-like protein, it is not known if this protein is expressed. Our first objective, therefore, was to investigate whether the V protein was expressed from the HPIV3 genome. To achieve this, we created a recombinant (r)HPIV3 clone with the nucleotide sequence for a hexahistidine tag on the 5′ end of the P gene. This tag enabled the detection of any protein that might be expressed from the start codon of the P gene. We speculated that if a V-like protein was not to be expressed, then the PD protein might be expressed in lieu of the V protein, which might result in no interference with the primary antiviral response by inhibition of IFN-β expression. However, it might also be possible that the D domain of the PD protein has a dual function and might facilitate viral RNA synthesis as well. For example, the HPIV3 C protein has been shown to function in two ways: it inhibits viral mRNA transcription, which leads to attenuated virus replication. It also inhibits the phosphorylation of STAT1 (signal transducer and activator of transcription 1), which blocks secondary IFN-β signaling [11, 12]. Accordingly, our second objective was to identify a possible function of the D domain of the PD protein in the HPIV3 replication cycle. We, therefore, constructed knockout viruses deficient in the expression of the C-terminal domain. We analyzed replication kinetics of the knockout viruses in permissive MA-104 and semi-permissive A549 cell types and examined the cytokine profile of infected A549 cells.
Materials and methods
Cells and viruses
Embryonic African green monkey kidney cells (MA-104, Clone 1, CRL-2378.1) were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained at 37 °C and 5 % CO2 in minimal essential medium (MEM), supplemented with 10 % fetal bovine serum (FBS), both purchased from Hyclone, Fisher Scientific, UT. Human lung epithelial carcinoma cells (A549, CCL-185) were also obtained from the ATCC and maintained under identical conditions with the exception of Dulbecco’s Modified Eagle’s Medium as the growth medium (DMEM, Hyclone). The wild-type (WT) HPIV3 virus, isolate 14702, was provided by Dr. J. Bouvin at St. Justine Hospital in Montreal, Canada, and propagated in MA-104 cells. A recombinant vaccinia virus that expresses the bacteriophage T7 RNA polymerase, vTF7-3, was generously provided by Dr. Bernard Moss [13]. During all assays, MA-104 and A549 cells were incubated in MEM supplemented with 2 % fetal bovine serum (FBS, Hyclone) and 50 μg/ml gentamicin (Sigma, St. Louis, MO).
Plasmid construction
The nucleotide sequence that encodes six histidine residues was inserted into the recombinant rHPIV3 antigenome immediately following the start codon of the P gene using the QuikChange® XL Site-Directed Mutagenesis Kit (Agilent Stratagene, Clara, CA) and the 1–5,267 cDNA segment [7, 14]. The forward primer, 5′-CTCAATCAATAGAGAGTTGATGCATCACCATCACCATCACGAAAGCGATGCTAAAAACTATC-3′, and reverse primer, 5′-GATAGTTTTTAGCATCGCTTTCGTGATGGTGATGGTGATGCATCAACTCTCTATTGATTGAG-3′, were used in the reaction to insert the nucleotide sequence for the hexahistidine tag, underlined, at viral position 1,787. The insertion of the tag was confirmed by sequencing, and the resulting clone was named pUC19-NT. Two additional clones were also engineered to eliminate the P gene editing site and silence the D domain of the PD protein using pUC19-NT as the template and QuikChange® XL Site-Directed Mutagenesis Kit (Agilent Stratagene). The forward primer, 5′-CTTCAACACATCAAGAAGATGACAAAAGAGGGAAAGACTGGTTT-3′, and reverse primer, 5′-AAACCAGTCTTTCCCTCTTTTGTCATCTTCTTGATGTGTTGAAG-3′, were used to delete 18 nucleotides at viral positions 2,495–2,512 from the antigenome. The deletion was confirmed by sequencing, and the resulting clone was named pUC19-DS. Also, the forward primer, 5′-CAAAAGAATTAAAAAAGGGGGAAAAGTGAAAGACTGGTTTAAGAAATCAAAAG-3′, and reverse primer, 5′-CTTTTGATTTCTTAAACCAGTCTTTCACTTTTCCCCCTTTTTTAATTCTTTTG-3′, were used to mutate a single nucleotide from G to T, underlined, at viral position 2,514. The deletion was confirmed by sequencing, and the resulting clone was named pUC19-SD. The assembly of the complete antigenomes for each mutant clone was performed as previously described [7, 14] and were renamed: pUC19-L which contained the hexahistidine tag and no P gene mutations, pUC19-V which destroyed the editing site, and pUC19-X which silenced the D domain of PD protein.
Rescue of infectious virus from cDNA
The procedure to rescue infectious, recombinant HPIV3 viruses from cDNA clones was adapted with modifications from Roth et al. [7, 14]. MA-104 cells were used as the host cell to infect vTF7-3 and transfect the three support plasmids, 1 μg of pTNT-NP, 2 μg of pTNT-P, and 0.05 μg of pTNT-L, and 0.5 μg of either of the genomic plasmids, pUC19-L, pUC19-V, or pUC19-X. The rescued recombinant viruses, rHPIV3-NT by pUC19-L transfection, rHPIV3-ΔES by pUC19-V transfection, and rHPIV3-ΔD by pUC19-X transfection, were amplified a final time in MA-104 cells for 3 days at 37 °C. The infected cells were scraped from the tissue culture wells and the cell lysates were frozen at −80 °C until used in each experiment. Viral genomic RNA was purified as previously described [7, 14] and sequenced to identify mutations contained in the P gene using SuperScript® III First-Strand Synthesis Supermix (Invitrogen™, Life Technologies, Grand Island, NY) and the 5′-GCAACACCAGATGATGAAGAAGA-3′ forward primer for cDNA synthesis. The cDNA products were amplified using AccuPrime™ Pfx SuperMix (Invitrogen™), the forward primer mentioned above, and the 5′-GGATTACACCAAGATTCTGCAATAG-3′ reverse primer. Amplification products were then sequenced.
Western blot analysis
Twelve-well plates were seeded with MA-104 cells and infected at an MOI of 0.1 with 3 × 104 PFU of the following viruses separately: rHPIV3-NT, rHPIV3-ΔES, or rHPIV3-ΔD. Each virus was absorbed to the cells for 2 h at 37 °C, after which the virus inocula were replaced with fresh MEM supplemented with 2 % FBS. The plates were incubated at 37 °C for 2 days. Protein from each infection was extracted using Mammalian Protein Extraction Reagent (M-PER®), and concentrations were measured using the colorimetric BCA protein assay kit (both from Pierce, Thermo Fisher Scientific, Rockford, IL). Absorbance measurements at 562 nm were taken with a SpectraMax® Plus384 spectrophotometer and data were analyzed with SoftMmax® Pro software, version 4.3 (both from Molecular Devices, Sunnyvale, CA). Twenty μg of each protein sample was mixed with NuPAGE® LDS Sample Buffer and sample reducing agent, heated at 70 °C for 10 min, loaded on a NuPAGE® Novex® 4–12 % Bis–Tris Gel, and electrophoresed at 200 V for approximately 1 h in NuPAGE® MOPS SDS Running Buffer and antioxidant (all reagents from Invitrogen™). The proteins in the gel were transferred to nitrocellulose using the iBlot™ 7-Minute Blotting System (Invitrogen™). The unbound protein binding sites on the nitrocellulose membrane were bound with SuperBlock® T20 (TBS) Blocking Buffer (Pierce) for 1 h with rocking at room temperature. Hexahistidine-tagged proteins were detected by the following chemiluminescent procedure. The monoclonal anti-polyhistidine-peroxidase antibody (Clone HIS-1, Sigma, St. Louis, MO), 4 μg/ml, was incubated with the nitrocellulose membrane for 1 h with rocking at room temperature. The membrane was washed three times with tris-buffered saline (TBS) containing 0.05 % Tween-20 (Pierce) for 5 min with rocking at room temperature. The antibody-bound proteins were then incubated with the SuperSignal® West Pico Chemiluminescent Substrate (Pierce) to detect any chemiluminescence. The membrane was photographed with the Image Station 2000R camera using a 1-min exposure, and the image was analyzed with the 1D image analysis software (both from Eastman Kodak Company). The 1D image analysis software was also used to estimate the molecular mass of each band in each lane by comparison to the BenchMark™ His-tagged Protein Standard (Invitrogen™).
MA-104 and A549 replication plaque assays
To assess the titers of HPIV3 WT, rHPIV3-NT, rHPIV3-ΔES, and rHPIV3-ΔD viruses, the plaque assay procedure was used [7]. Based on results from this assay, virus inocula were calculated and adjusted to normalize the PFU (plaque forming units) of each virus used in all subsequent experiments. Titers for the four above-mentioned viruses were also assessed in A549 cells. For each titration, duplicate 12-well plates were seeded with A549 cells, and cells were infected at an MOI of 0.1 with 4.8 × 104 PFU of one of the four viruses. The virus inoculum was absorbed to the cells for 2 h at 37 °C. The virus was then removed and replaced with fresh MEM supplemented with 2 % FBS. Plates were incubated at 37 °C for 3 days after which infected A549 cells were scraped from the tissue culture wells and virus lysates were frozen at −80 °C. Each virus was plaque-titered in triplicates.
MA-104 and A549 one-step replication curves
For each virus titered, duplicate 12-well plates were seeded with MA-104 or A549 cells and infected at an MOI of 3 with either 9.1 × 105 or 1.4 × 106 PFU. Each of the following viruses were titered: rHPIV3-NT, rHPIV3-ΔES, and rHPIV3-ΔD. Virus was absorbed to the cells for 2 h at 37 °C, removed, replaced with fresh MEM supplemented with 2 % FBS, and cells were incubated at 37 °C. Infected cells were scraped from individual tissue culture wells at 2, 12, 24, and 48 h post infection and frozen at −80 °C. Each time point for each virus was quantified by plaque assay in triplicate as described earlier.
MA-104 viral qRT-PCR assay
To assess viral genomic RNA replication and mRNA transcription, RNA from rHPIV3-NT, rHPIV3-ΔES, and rHPIV3-ΔD viral infections was analyzed by a procedure previously described [7].
A549 cytokine ELISA
For each virus, triplicate 24-well plates were seeded with A549 cells and infected at an MOI of 0.1 with 3.5 × 104 PFU and the effects of rHPIV3-NT, rHPIV3-ΔES, and rHPIV3-ΔD viruses on cytokine production in A549 cells were assessed. Each virus was absorbed for 2 h at 37 °C followed by virus replacement with fresh MEM supplemented with 2 % FBS, and an incubation period at 37 °C for 24 h. Tissue culture medium for the infected and uninfected wells was removed and stored at −80 °C. Cellular RNA was isolated from the monolayer from the same tissue culture wells and used for quantitative real-time polymerase chain reaction (qRT-PCR), as discussed below. Cytokine levels present in the tissue culture media were measured by enzyme-linked immunosorbent assay (ELISA) using the VeriKine™ Human Interferon Beta ELISA Kit (PBL InterferonSource, Piscataway, NJ). Absorbance was measured on a SpectraMax® Plus384 spectrophotometer at 450 nm, and data were analyzed with SoftMax® Pro software. IFN-β expressed from MA-104 cells infected with 2.5 × 104 PFU, MOI of 0.1, of rHPIV3-NT was also assayed using the same procedure and ELISA kit as described above. A human multiplex cytokine/chemokine array was used following the manufacturer’s protocol (Cytokine Research Institute) to measure the expression levels of the following proteins also present in the same supernatants: interleukin (IL)-1α, IL-6, IL-8, IL-10, IL-12p70, monocyte chemotactic protein 1 (MCP-1), macrophage inhibitory protein 1α (MIP-1α), and MIP-1β, regulated and normal T cell expressed and secreted (RANTES), gamma interferon (IFN-γ), tumor necrosis factor alpha (TNFα), and eotaxin (EO). Chemiluminescence was measured with a Quansys Imager, and data were analyzed with Q-View™ software, version 2.0 (both from Quansys Biosciences, Logan, UT). Concentrations of cytokines and chemokines present in the tissue culture media were determined either by linear or exponential regression analysis. Concentrations of each cytokine and chemokine expressed from A549 infected and uninfected cells were compared for significance using unpaired, two-tailed Student’s t tests (Microsoft Excel).
A549 IFN-β qRT-PCR assay
The CellsDirect™ One-Step qRT-PCR Kit (Invitrogen™, Life Sciences, Grand Island, NY) was used to amplify mRNA encoding the IFN-β and β-actin genes from total RNA isolated from infected and uninfected A549 cells 24 h after virus exposure. Following DNase I treatment of the total RNA samples, one quarter of each sample was added to duplicate wells of hard-shell 96-well skirted PCR plates. FAM-labeled forward primer (200 nM) and unlabeled reverse primer (250 nM) for the IFN-β D-LUX™ Select Gene Expression Assay (Invitrogen™) along with JOE-labeled forward primer (100 nM) and unlabeled reverse primer (125 nM) for the β-actin Certified LUX™ Primer Set (Invitrogen™) were added to the qRT-PCR mixture. Also, an additional one unit of Platinum® Taq DNA Polymerase (Invitrogen™) and 3 mM MgSO4 was added to the qRT-PCR mixture. The reaction mixtures were incubated in a DNA Engine Opticon 2 real-time PCR detection system (BioRad, Hercules, CA). Opticon Monitor™ software was used to calculate the relative expression of 2^(-delta delta Ct) of IFN-β during each infection using β-actin as an internal control and the IFN-β expressed in uninfected cells as the baseline calibrator [15]. The relative expression levels of IFN-β from cells infected with each virus were compared and statistically analyzed using unpaired, two-tailed Student’s t tests.
Results
Rescue of infectious, recombinant HPIV3 viruses expressing tagged proteins from the P gene
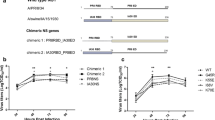
Nucleotides that encode for a hexahistidine (6x-His) tag were inserted into the P gene of the rHPIV3-NT parent clone immediately downstream of the start codon. This addition created a tag on all proteins expressed from the P gene start codon, including the known P and PD proteins and possibly an unidentified, putative W protein or theoretical V protein. Eighteen total nucleotides were added to the viral antigenome to conform to the “Rule of Six,” which is necessary for optimal paramyxovirus replication [16, 17]. Two other knockout clones, also containing the 6x-His tag, were made to silence the expression of the D domain of the PD protein in two different fashions. In the first approach, 18 nucleotides, following the “Rule of Six,” were removed from the editing site within the P gene of the rHPIV3-NT antigenome, which did not remove or interfere with known P protein domains, and the rescued clone was named rHPIV3-ΔES. In the second approach, a single nucleotide was substituted to create a stop codon that silenced only the D domain of the PD protein, which did not interfere with the expression of the P or W protein, and the resulting clone was named rHPIV3-ΔD. Sequence analysis of the three rescued rHPIV3 viruses showed an intact editing site in the rHPIV3-NT clone, the elimination of the editing site in the rHPIV3-ΔES clone, and a single substitution mutation in the rHPIV3-ΔD clone (Fig. 1a).
Nucleotide sequence and Western blot analysis of recombinant HPIV3 viruses. a The cDNA clone of the parental (wild-type-like) virus, rHPIV3-NT, was mutated to create two clones where the expression of the D domain of the PD protein was silenced. In the first mutant cDNA clone, nucleotides 2,495–2,512 were removed from the genomic sequence of rHPIV3-NT to eliminate the editing site (ES) and create the rHPIV3-ΔES knockout virus. In the second mutant cDNA clone, nucleotide 2,514 was mutated from G to T, a nonsense mutation, to create the rHPIV3-ΔD knockout virus. b Western blot analysis of MA-104-infected cell lysates of rHPIV3-NT, rHPIV3-ΔES, and rHPIV3-ΔD. The nucleotide sequence that encodes a hexahistidine (6xHis) tag was inserted immediately downstream of the start codon of the phosphoprotein P in all three recombinant HPIV3 viruses, rHPIV3-NT, rHPIV3-ΔES, and rHPIV3-ΔD, marked by an asterisk below the schematic representation of the viral antigenome. Proteins from MA-104 cell lysates infected with each virus were separated by SDS-PAGE and blotted onto nitrocellulose. 6xHis-tagged proteins were probed with a monoclonal anti-6xHis antibody conjugated to peroxidase and detected using chemiluminescence. Arrows indicate positions of 6xHis-tagged viral proteins: the P protein at ~78 kDa, the PD protein at ~50 kDa, and the W protein at ~36 kDa
Proteins expressed from the P gene of the three viruses were detected with anti-6xHis antibodies, which bind to the 6x-His tag on the N-terminal end. Western blot analysis of infected cell lysates with the wild-type-like recombinant HPIV3 virus (rHPIV3-NT, no deletions) identified the P protein at approximately 78 kilo Daltons (kDa), the PD protein at ~50 kDa, and the W protein at ~36 kDa (Fig. 1b). No other proteins translated from the P gene start codon, including the V protein, were visible. In cell lysates infected with the recombinant HPIV3 virus missing the editing site (rHPIV3-ΔES), only the P protein was detected but not the bands representing the W and PD proteins (Fig. 1b). The dramatic increase in the intensity of the P protein band might be attributed to the absence of an active editing site, and, therefore, all P mRNAs were translated into one P protein rather than three distinct proteins. Finally, in cell lysates infected with recombinant HPIV3 virus with the D domain silenced (rHPIV3-ΔD), only the P and W protein, but not the PD or W protein were detected (Fig. 1b). The C protein, which is expressed from an alternative reading frame, was not visible in our system because it did not contain the 6xHis tag.
Replication of wild-type and recombinant HPIV3 viruses in MA-104 and A549 cells
To compare the replication kinetics of four HPIV3 viruses (HPIV3 WT, wild-type strain; rHPIV3-NT, recombinant parental strain; rHPIV3-ΔES and rHPIV3-ΔD, two recombinant knockout strains), each virus was amplified in MA-104 and A549 cells separately, and titers were determined by plaque assay. In both MA-104 and A549 cells, the titers of the three recombinant viruses were comparable to those of the wild-type and parental strains with the exception of rHPIV3-ΔES in A549 cells (Table 1). Similar titers between HPIV3 WT and rHPIV3-NT suggest that the addition of the 6x-His tag did not interfere with virus replication as seen with other 6x-His-tagged viral proteins, such as flavivirus NS2B/NS3 proteases and herpes simplex virus ICP27/ICP8 late gene transcription proteins [18, 19]. The rHPIV3-ΔES virus, which has 18 nucleotides encompassing the editing site removed, had significantly reduced titers compared to wild-type and parental viral strains in A549 cells (Table 1, p < 0.01). In addition, the growth characteristics of the three recombinant viruses were compared by infecting MA-104 and A549 cells with equivalent multiplicities of infection (MOI) and assaying virus titers at different time points during the infection. In both cell lines, the replication curves for the rHPIV3-ΔES and rHPIV3-ΔD knockout viruses were slightly reduced, yet comparable to the replication curve for the parental virus rHPIV3-NT (Fig. 2a, b).
Growth kinetics of recombinant HPIV3 viruses in MA-104 cells and A549 cells. a Permissive MA-104 cells and b semi-permissive A549 cells were infected with the parental virus rHPIV3-NT (unfilled circle), or the knock-out viruses rHPIV3-ΔES (filled square) and rHPIV3-ΔD (filled triangle). Cells were harvested at the indicated times post infection, and the amount of virus present in the culture medium was determined by plaque assay. Data represent the mean ± SD of two independent experiments
Viral genomic replication and gene transcription of the recombinant HPIV3 viruses in MA-104 cells
To investigate if the D domain of the PD protein may influence viral RNA synthesis, viral genomic replication and gene transcription of each recombinant virus were measured by qRT-PCR at various time points after infection of MA-104 cells. Two different primers and an oligo(dT)20 primer were used to prime viral RNA and to differentiate between viral gene transcription and genomic replication. These were gene-specific primers that only bind to negative-sense genomic viral RNA. In both cases, viral gene transcription and genomic replication were significantly reduced in both knockout viruses (rHPIV3-ΔES and rHPIV3-ΔD) when compared to the different viral RNA synthesis processes of the parental virus, rHPIV3-NT (Fig. 3a, b, p < 0.01). These results suggest that the D domain of the PD protein might be involved in the regulation of viral RNA synthesis.
RNA synthesis of recombinant HPIV3 viruses. Relative expression differences of viral mRNA transcription (a) and genomic replication (b) of the parental virus, rHPIV3-NT (unfilled circle), and knockout viruses, rHPIV3-ΔES (filled square) and rHPIV3-ΔD (filled triangle) were measured by qRT-PCR. At specific time points, RNA was extracted from infected MA-104 cells and primed for reverse transcription to detect specific populations of viral RNA. To measure viral genomic replication, viral genomic RNA was primed with an HPIV3-specific primer, which straddles the intergenic region between the fusion and hemagglutinin-neuraminidase genes, and only anneals to viral, negative-sense, genomic RNA. To measure viral transcription, viral mRNA was primed with an oligo(dT)20 primer. Each set of cDNAs was PCR-amplified with HPIV3-specific primers that were tagged with FAM. 2^(delta Ct) relative expression differences were calculated for each virus at each time point, using the 0 h measurement for each virus as the calibrator. Significant differences were seen for both knockout viruses when compared to the parental virus (p < 0.01). Data represent the mean ± S.D. of triplicate assays
Cytokine expression profiles from A549 cells infected with the recombinant HPIV3 viruses
To investigate if the D domain of the PD protein influences the expression of IFN-β or other cytokines, cytokine/chemokine levels excreted from A549 cells infected with each recombinant virus were assayed. Preliminary time course evaluation of IFN-β expressed from A549 cells infected with the rHPIV3-NT virus showed peak expression at 24 h (data not shown). Therefore, cytokine expression studies were measured only at this time point to conserve resources. Of the cytokines measured by ELISA, only the expression levels of five cytokines/chemokines were significantly higher in infected cells when compared to the cytokine/chemokine levels in uninfected cell controls (Fig. 4, p < 0.01). These included the chemokine RANTES (regulated upon activation, normal T cell expressed and secreted), and the cytokines IL-6 (interleukin 6), IL-8, MCP-1 (monocyte chemotactic protein 1), and IFN-β (Fig. 5). Expression levels of RANTES, IL-6 and IFN-β were also significantly decreased in cells infected with both knockout viruses when compared to cells infected with the parental strain virus (Figs. 4a, b, 5a, p < 0.01). On the other hand, the expression levels of IL-8 and MCP-1 secreted from A549 cells infected with all three recombinant viruses separately were similar (Fig. 4c, d). Expression levels of the RANTES chemokine and IFN-β cytokine expressed from A549 cells infected with the rHPIV3-ΔES virus were significantly increased compared to RANTES and IFN-β expressed from cells infected with the rHPIV3-ΔD virus (Figs. 4a, 5a, p < 0.01). To confirm the reduction in IFN-β expression levels in A549 cells infected with the knockout viruses, IFN-β mRNA levels were measured by multiplex qRT-PCR. IFN-β mRNA transcription levels were significantly lower in A549 cells infected with either knockout virus than those from cells infected with the parental virus (Fig. 5b, p < 0.05).
Cytokine expression profile of infected A549 cells as measured by ELISA. Plates were seeded with A549 cells and infected with each virus (NT, rHPIV3-NT; ΔES, rHPIV3-ΔES; ΔD, rHPIV3-ΔD). At 24 hpi (hours post infection), cell culture medium was removed and assayed for cytokine/chemokine expression by multiplex ELISA and compared to uninfected cell controls (mock). Of the 16 cytokines and chemokines measured, only RANTES (a), IL-6 (b), IL-8 (c), and MCP-1 (d) expressed from infected A549 cells resulted in significant increases compared to uninfected cell controls (p < 0.01) and are shown here. Results represent the mean ± SD of triplicate assays. Pairwise comparisons are denoted with a letter code
Beta interferon (IFN-β) expression and mRNA transcription of infected A549 cells as measured by ELISA and qRT-PCR. Plates were seeded with A549 cells and infected with each virus (NT, rHPIV3-NT; ΔES, rHPIV3-ΔES; ΔD, rHPIV3-ΔD). At 24 hpi (hours post infection), cell culture medium was removed and assayed for IFN-β expression by ELISA (a) and compared to uninfected cell controls (mock). In addition, RNA was extracted from the same infected A549 cells and IFN-β mRNA was measured by qRT-PCR (b). Cellular mRNA was primed with an Oligo(dT)20 primer and PCR-amplified using IFN-β/FAM and β-actin/JOE primers in a multiplex fashion. 2^(-delta delta Ct) relative expression differences were calculated for each virus using β-actin as an internal control and uninfected cell controls as the baseline calibrator. Results represent the mean ± S.D. of triplicate assays. Pairwise comparisons are denoted with a letter code. Means with the same letter are not significantly different from each other with p < 0.01 in (a) and p < 0.05 in (b), respectively
Discussion
During the replication of most paramyxoviruses, additional proteins are expressed from the multicistronic P gene by insertion of nontemplated G residues at a specific editing site. This RNA editing leads to a translational frame shift and the generation of chimeric polypeptides with the same N-terminus but different C-termini. In this study, the P, PD, and W proteins were the only proteins detected by Western blot analysis; the V protein was not detected. However, the HPIV3 V protein could still have been expressed at low levels using a different initiation codon either by internal ribosome binding sites, which are present in picornaviruses and hepatitis C viruses, or by ribosomal shunting, exhibited by adenoviruses and other paramyxoviruses [20–23].
In this study, we also found that in the absence of the D domain of the PD protein, the rHPIV3-ΔD and rHPIV3-ΔES viruses replicated at similar levels but showed reduced viral RNA synthesis, and reduced cytokine/chemokine expression from infected A549 cells when compared to the wild-type virus. This suggests that the mutated viruses were slightly attenuated. In a previous study, Durbin et al. [5] also silenced the D and theoretical V domains. Although viral RNA synthesis or cytokine expression levels were not determined in that study, significant reductions in viral titers of the knockout viruses were only found in lung tissue if both domains were silenced simultaneously in vitro. In the same study, the authors introduced three stop codons into the nucleotide sequence that encodes the D protein at amino acid (aa) positions 304, 305, and 306 to silence expression in the D domain of the PD protein and the V domain. In our study, only one stop codon was introduced into the nucleotide sequence that encodes the PD protein at aa position 245 which immediately follows the editing site located within the P gene. Recently, Wells and Malur [4] identified three nuclear localization signals in the PD protein, located at aa positions 225–241, 266–272, and 340–346. Therefore, the knockout viruses constructed by Durbin et al. [5] eliminated the last nuclear localization signal, whereas our knockout viruses silenced the last two signals. From the results of these three studies, it can be concluded that the latter two nuclear localization signals may promote more efficient HPIV3 RNA synthesis. To gain insight into any possible biologically relevant function of the D domain of the PD protein, results from our study need to be confirmed by repeating the experiments using in vivo models, such as Guinea pigs and cotton rats [24, 25].
Wells and Malur [4] also detected the PD protein in vivo and showed that it was localized primarily to the nucleus of infected HeLa cells. Leptomycin B treatment, which inhibits nuclear export mechanisms, led to retention of the PD protein in the nucleus. Therefore, the PD protein may act as a shuttle by recruiting host nuclear proteins or other cellular factors into the cytoplasm and or at least, assisting in some aspect of virus replication. Our results indicate that HPIV3 viral RNA synthesis is adversely affected during the expression of a PD protein minus the D domain. Thus, perhaps the HPIV3 PD protein recruits a cellular nuclear protein that aids in viral RNA synthesis, such as has been shown for the RNA-binding La protein [26]. The La protein was found to interact with the viral RNA of various single-stranded RNA viruses, including HPIV3, and contribute to virus replication [27]. In addition, during RNAi-La protein knockdown experiments, the absence of the La protein resulted in attenuated replication of respiratory syncytial virus (RSV), another paramyxovirus, in an IFN-independent manner [28]. These studies demonstrate that the La protein is intimately involved in virus replication and may facilitate viral RNA synthesis directly. Further studies, such as two-hybrid experiments, are needed to determine if in fact the HPIV3 PD protein interacts with the cellular La protein during viral RNA synthesis.
In our study, we also found that the elimination of the P gene’s editing signal and site in the rHPIV3-ΔES virus resulted in significantly reduced viral replication and significantly increased levels of IFN-β and RANTES expression in infected A549 cells compared to the rHPIV3-ΔD virus. These results imply that the rHPIV3-ΔES virus is a significantly attenuated virus because it seems to have induced a stronger cellular antiviral response in the form of IFN-β levels. Six aas, IKKGGK, at aa positions 238–243 were deleted from the P protein of the rHPIV3-ΔES virus that coincides with a portion of the third nuclear localization signal, aa positions 225–241, identified by Wells and Malur [4]. Since A549 cells expressed IFN-β, thus exhibiting an antiviral response, the alteration of the third nuclear localization signal in the rHPIV3-ΔES virus and subsequent differences in viral replication and cellular cytokine levels suggests that this signal may play an important role in counteracting the host cell’s antiviral response. This third signal is located just upstream from the editing site in the P gene and is, therefore, located on all three proteins expressed from the start codon of the P gene, namely the P, PD, and W protein. Therefore, maybe all three P gene-associated proteins influence the host cell’s antiviral response using this signal. Alternatively, this is perhaps a main function of the W protein since the W protein is truncated soon after the editing site and is expressed in higher concentrations than the P and PD proteins.
In summary, our results suggest that the V protein most likely is not expressed in cell culture, the D domain of the HPIV3 PD protein may facilitate viral RNA synthesis in a luxury manner, and that a nuclear localization signal located on the P, PD, and W protein may be involved in counteracting the host cell’s antiviral response by regulating the IFN-β expression. Further studies are needed to confirm these results and elucidate the specific functions of these domains and signals.
References
M.S. Galinski, R.M. Troy, A.K. Banerjee, Virology 186, 543–550 (1992)
S. Vidal, J. Curran, D. Kolakofsky, J. Virol. 64, 239–246 (1990)
Y. Matsuoka, J. Curran, T. Pelet, D. Kolakofsky, R. Ray, R.W. Compans, J. Virol. 65, 3406–3410 (1991)
G. Wells, A. Malur, Virus Genes 37, 358–367 (2008)
A.P. Durbin, J.M. McAuliffe, P.L. Collins, B.R. Murphy, Virology 261, 319–330 (1999)
R.G. Paterson, G.P. Leser, M.A. Shaughnessy, R.A. Lamb, Virology 208, 121–131 (1995)
J.P. Roth, J.K. Li, D.F. Smee, J.D. Morrey, D.L. Barnard, Antiviral Res. 82, 12–21 (2009)
C.K. Pfaller, K.K. Conzelmann, J. Virol. 82, 12365–12373 (2008)
J. Andrejeva, K.S. Childs, D.F. Young, T.S. Carlos, N. Stock, S. Goodbourn, R.E. Randall, Proc Natl Acad Sci USA 101, 17264–17269 (2004)
K. Childs, N. Stock, C. Ross, J. Andrejeva, L. Hilton, M. Skinner, R. Randall, S. Goodbourn, Virology 359, 190–200 (2007)
A.G. Malur, S. Chattopadhyay, R.K. Maitra, A.K. Banerjee, J. Virol. 79, 7877–7882 (2005)
A.G. Malur, M.A. Hoffman, A.K. Banerjee, Virus Res. 99, 199–204 (2004)
T.R. Fuerst, E.G. Niles, F.W. Studier, B. Moss, Proc Natl Acad Sci USA 83, 8122–8126 (1986)
Roth J.P., Li J.K., and Barnard D.L., Curr Protoc Microbiol Chapter 15, Unit 15F.1 (2010)
M.W. Pfaffl, Nucleic Acids Res. 29, 2002–2007 (2001)
P. Calain, L. Roux, J. Virol. 67, 4822–4830 (1993)
A.P. Durbin, J.W. Siew, B.R. Murphy, P.L. Collins, Virology 234, 74–83 (1997)
M. Olesky, E.E. McNamee, C. Zhou, T.J. Taylor, D.M. Knipe, Virology 331, 94–105 (2005)
M. Bessaud, B.A. Pastorino, C.N. Peyrefitte, D. Rolland, M. Grandadam, H.J. Tolou, Virus Res. 120, 79–90 (2006)
D. Boehringer, R. Thermann, A. Ostareck-Lederer, J.D. Lewis, H. Stark, Structure 13, 1695–1706 (2005)
C. Junemann, Y. Song, G. Bassili, D. Goergen, J. Henke, M. Niepmann, J. Biol. Chem. 282, 132–141 (2007)
Q. Xi, R. Cuesta, R. Schneider, J. Virol. 79, 5676–5683 (2005)
P. Latorre, D. Kolakofsky, J. Curran, Mol. Cell. Biol. 18, 5021–5031 (1998)
E.M. Kudlacz, L.E. Baugh, W.P. Porter, M.T. Kenny, A.M. Farrell, Lab. Anim. Sci. 43, 445–453 (1993)
T.F. Murphy, E.J. Dubovi, W.A. Clyde, Exp. Lung Res. 2, 97–109 (1981)
J. Rinke, J.A. Steitz, Cell 29, 149–159 (1982)
B.P. De, S. Gupta, H. Zhao, J.A. Drazba, A.K. Banerjee, J. Biol. Chem. 271, 24728–24735 (1996)
V. Bitko, A. Musiyenko, M.A. Bayfield, R.J. Maraia, S. Barik, J. Virol. 82, 7977–7987 (2008)
Acknowledgments
We would like to thank Dr. Steven Aust, Dr. Thomas Bunch, and Dr. Donald Smee for their review of the manuscript. We would also like to thank Dr. Craig Day, Dr. Brian Gowen, and Tyler Mclean for their technical assistance. This work was supported by Contract No. N01 AI-30048 from the Virology Branch of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). Joseph K.-K. Li was also supported in parts by AES Project UTA 01050.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roth, J.P., Li, J.KK., Morrey, J.D. et al. Deletion of the D domain of the human parainfluenza virus type 3 (HPIV3) PD protein results in decreased viral RNA synthesis and beta interferon (IFN-β) expression. Virus Genes 47, 10–19 (2013). https://doi.org/10.1007/s11262-013-0919-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-013-0919-x