Abstract
The P gene of human parainfluenza virus type 3 (HPIV 3) encodes a multicistronic P mRNA that gives rise to four polypeptides. The P and C proteins are synthesized from two discrete overlapping AUG codons from the unedited P mRNA, while synthesis of two additional proteins, V and PD, presumably occurs via a unique transcriptional editing mechanism. However, the presence of V and PD proteins in HPIV 3 infected cells and their role in viral replication remains uncertain. Here, in vitro expression of full-length PD protein from an altered P mRNA and generation of a polyclonal antibody to the COOH-terminus of PD was achieved. Confocal immunofluorescence analysis following Leptomycin B (LMB) treatment revealed the presence of PD protein in nuclear and cytoplasmic compartments of HPIV 3 infected cells suggesting the involvement of a nuclear localization signal in this process. These initial results provide new impetus for further characterization of the role of PD in HPIV 3 infection.
Similar content being viewed by others
Introduction
Human parainfluenza virus type 3 (HPIV 3) is the causative agent of bronchiolitis, pneumonia, and croup, and is a member of the genus Respirovirus within the subfamily Paramyxovirinae [1, 2]. The non-segmented, negative sense RNA genome comprising 15,462 nucleotides is arranged co-linearly in the order 3′-[leader (le)-N-P-M-F-HN-L-trailer (tr)]-5′ and encapsidated by the nucleoprotein (N) to form a helical nucleocapsid, i.e. the N-RNA, which serves as a template for the synthesis of mRNAs during transcription and for the synthesis of full-length copy of the genome RNA during replication [3]. The functional viral RNA-dependent RNA polymerase composed of the large, multifunctional protein (L) and a phosphoprotein (P) together with host factors initiates transcription of the viral genome and synthesizes five monocistronic mRNAs that subsequently give rise to N, M, F, HN, and L proteins [4–6].
However, as with other members within this subfamily, the HPIV 3 P gene gives rise to a unique P mRNA that is capable of synthesizing several polypeptides [1, 2]. A major polypeptide that arises from the unedited P mRNA encodes the phosphoprotein P (603 amino acids), an oligomeric protein that is phosphorylated both in vitro and in vivo [7–9] and forms a stable P–L complex to enhance the processivity of L during transcription [8, 10]. In addition, P also interacts with N protein to form a soluble P–N complex which is essential for the encapsidation of nascent RNA chains during replication [11].
The generation of a second polypeptide from within the P mRNA occurs via the utilization of an alternative AUG codon in the +1 open reading frame (ORF) relative to the AUG codon of P mRNA and leads to the synthesis of a basic polypeptide, the C protein (199 amino acids) (Fig. 1), which shares a number of features common to other Paramyxovirinae C proteins [12–14], including binding to its cognate L protein in a homologous interaction leading to subsequent down regulation of viral mRNA synthesis in vitro and in vivo [15–22] as well as interacting in a heterologous manner with Sendai virus polymerase complex to inhibit viral replication [21, 23]. Furthermore, as with other Paramyxovirinae C proteins, the HPIV 3 C protein is also capable of abrogating the host interferon signaling pathway by specifically inhibiting the phosphorylation of key intermediary elements of the JAK-STAT pathway [24–27] as well as attenuation of HPIV 3 replication both in vitro and in vivo [28].
(a). Organization of HPIV 3 P mRNA and generation of P, PD, and C ORF’s. The initiation (AUG) and termination codons (TAA, TAG) are numbered according to the complete genomic sequence, P ORF: 1784–3595, PD ORF: 1784–2903, and C ORF: 1795–2393. The insertion of two extra G residues within the “editing site” of P mRNA at position 2509 followed by a fusion of D ORF (black box) with the upstream P ORF is shown. The 3′ gene start (GE) and 5′ gene end (GE) sequences are denoted by dotted lines. The sizes of P (603 aa), PD (372 aa), and C (199 aa) are indicated. Figure not drawn to scale. (b). Expression of HPIV 3 PD in vitro. Plasmids pGEM 4 and pGEM 4 PDflag were transcribed in rabbit reticulocytes in vitro using the TNT System (Promega) in the presence of [35S]-methionine and labeled polypeptide was separated by SDS-PAGE followed by autoradiography. The position of PD is indicated by an arrow
Apart from P and C proteins, the HPIV 3 P mRNA is also capable of synthesizing two additional polypeptides, V and PD proteins, via a co-transcriptional editing mechanism that involves insertion of one or two G residues at an “editing site” within the P mRNA leading to the formation of V and PD proteins, respectively [29]. However, the insertion of a single G residue during this process leads to the generation of two translation stop codons immediately downstream of the “editing site” resulting in the premature termination of the V ORF [28, 29]. In contrast, addition of two G residues at the “editing sites” leads to the synthesis of a 131 amino acid long polypeptide from D ORF that is subsequently fused to the 241 amino acids encoded by the P ORF thus generating a chimeric PD protein (372 amino acids) (Fig. 1). The presence of both V and PD proteins has remained uncertain and obscure as no biochemical evidence indicating their existence has been documented. Interestingly, “reverse genetics” approaches have successfully led to the isolation of mutant viruses possessing individual mutations within the V and D ORF’s and a double mutant devoid of both V and D ORF’s [28, 30]. The mutant viruses, rVKO and rDKO, harboring individual mutations within the V and D ORF’s did not exhibit any adverse effect on HPIV 3 replication either in vitro or in vivo. However, the double mutant virus, rDVKO, devoid of both V and D ORF’s, was shown to be significantly attenuated in vivo suggesting that V and D proteins may play an important role in HPIV 3 replication in vitro, in vivo, or both [28].
The alteration in viral replication following mutations of V and D ORF’s and their suggestive involvement in this process coupled with a lack of previous data therefore highlights the need toward functional characterization of these proteins in HPIV 3 infection. Herein, we have performed an initial investigation to verify the presence of PD protein with the long-term goal to functionally characterize its role in HPIV 3 infection. As a preliminary step toward achieving our goal, we manipulated the HPIV 3 P mRNA in vitro to encode the PD ORF and expressed the full-length PD protein using an E. coli expression system. In addition, we generated a polyclonal antibody specific to the COOH-terminus of PD to facilitate the intracellular localization of PD following HPIV 3 infection. Results from double-labeled immunofluorescent confocal microscopy analysis revealed that PD is localized as a cytoplasmic and predominantly nuclear protein in HPIV 3 infected cells. This report provides first evidence for the intracellular localization of PD allowing us to pursue further studies to dissect the role of PD in the HPIV 3 life cycle.
Materials and methods
Cells and viruses
HeLa cells were cultured in Dulbecco’s modified medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), 100 U/ml penicillin (Invitrogen) and 100 U/ml streptomycin (Invitrogen), and 2 mM glutamine (Invitrogen). Cells were maintained at 37°C and 5% CO2. Recombinant vaccinia virus vTF7-3 that expresses the T7 RNA polymerase was grown in HeLa cells. HPIV 3 (HA-1, NIH 47885) was propagated on CV-1 cells as described previously [4, 5].
Plasmids and site-directed mutagenesis
The generation of the PD ORF from HPIV 3 P gene was achieved using a two step protocol. In the first step, a QuikChange mutagenesis (Stratagene) protocol involving two complementary oligonucleotides 5′ QC1PD-F and QC2PD-R (Table 1) was utilized for the insertion of two extra G residues within the editing site of HPIV 3 P mRNA of the plasmid pGEM 4 Pflag that harbors a FLAG epitope at the 3′ end of HPIV 3 P gene [10]. Following mutagenesis, two additional oligonucleotides PD-ER5F and PD-XHR with EcoR V and Xho I restriction sites, respectively (Table 1), were employed in a second amplification reaction to generate a 611 bp PCR fragment that was subsequently digested with EcoR V and Xho I enzymes and cloned into the corresponding sites of pGEM 4 Pflag to obtain the pGEM 4 PDflag plasmid.
In a different reaction, a DNA fragment encompassing 131 amino acids unique to the COOH-terminal of PD was amplified using oligonucleotides D-BMF and D-XHR (Table 1) and the resulting fragment with BamH I and Xho I restriction sites at the 5′ and 3′ extremities, respectively, was cloned into the corresponding sites of pGEM 4 Pflag to obtain pGEM 4 Dflag plasmid. Finally, dideoxy sequencing of pGEM 4 PDflag and pGEM 4 Dflag, plasmids was performed to verify their sequence integrity and expression of the full-length PD protein was additionally confirmed by an in vitro transcription–translation coupled system (Promega) using [35S]-methionine as the labeled precursor following the manufacturer’s protocol.
Vaccinia virus/T7 RNA polymerase-mediated transient expression of PDflag
Transient expression of PDflag was achieved using the vaccinia virus-T7 RNA polymerase-mediated system as described earlier [31]. Briefly, HeLa cells (3 × 105) in 6-well plate were infected with a recombinant vaccinia virus (vTF7-3) that expresses the T7 RNA polymerase gene at a multiplicity of infection (MOI) of 1. At 1 h post-infection, cells were individually transfected with 2 μg each of pGEM 4, Flag epitope tagged pGEM 4 Pflag and pGEM 4 PDflag plasmids, or non-tagged pGEM 4 P and pGEM 4 PD plasmids using Lipofectin (Invitrogen) in OptiMEM (Invitrogen) according to manufacturer’s instructions. At 5 h post-transfection, the mixture was replaced with DMEM supplemented with 10% FBS. At 24 h post-transfection, cells were washed once with PBS and lysed in 150 μl of lysis buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% NP40) containing protease inhibitor cocktail (Sigma) and cell lysates were resolved onto SDS-10% polyacrylamide gels.
Bacterial expression of full-length PD and the COOH-terminus D polypeptide
Plasmids pGEM 4 PDflag and pGEM 4 Dflag were digested with BamH I and Xho I enzymes and the resulting fragments were individually cloned into the corresponding BamH I and Xho I sites present downstream of the polyhistidine tag within the E. coli expression vector, pRSET-A (Invitrogen) to obtain pRSET-PD and pRSET-D plasmids, respectively. Following restriction analysis, plasmid DNA from two individual positive clones was transformed into E. coli BL21 (DE3) pLysS and small-scale cultures were analyzed for induction of the recombinant proteins. Cells were grown to an OD600 of 0.4–0.6 in LB broth containing 100 μg/ml ampicillin and protein expression was induced by addition of 1 mM IPTG followed by an additional 4 h at 37°C. Bacterial cells were harvested by centrifugation at 6,000g for 5 min at 4°C and resuspended in phosphate buffer (20 mM sodium phosphate, 0.5 M NaCl pH 7.5). The cell suspensions were kept on ice and disrupted by a French press. After centrifugation at 12,000g for 15 min at 4°C, the soluble supernatant fraction was collected and aliquots were analyzed by SDS-PAGE. Gels were either stained with Coomassie Brilliant Blue R 250 or processed for Western blot analysis.
Purification of D polypeptide and generation of affinity purified polyclonal antibody
A large-scale culture (200 ml) culture of E. coli BL21 (DE3) pLysS harboring the pRSET-D plasmid was grown at 37°C and induced with 1 mM IPTG and cells were harvested by centrifugation at 6,000g for 5 min at 4°C as described above. The bacterial pellet was resuspended in buffer A (8 M urea, 20 mM sodium phosphate, 0.5 M NaCl pH 8.0) followed by disruption of the cells using a French press. After centrifugation at 12,000g for 15 min at 4°C, the soluble supernatant fraction was applied onto a Ni-agarose affinity column (Invitrogen) previously equilibrated with buffer A. The column was washed with five volumes of buffer A followed by two successive washes with buffer B (8 M urea, 20 mM sodium phosphate, 0.5 M NaCl pH 6.3). The bound protein was finally eluted with two elution buffers containing 8 M urea, 20 mM sodium phosphate, 0.5 M NaCl over a pH gradient of 5.9 and 4.5, respectively. Aliquots of fractions containing the eluted protein were analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue R 250. Fractions containing the purified protein were pooled and desalted using a D-Salt column (Pierce). The protein concentration was measured by Bradford assay (Bio-Rad).
The purified D polypeptide was utilized for the generation of an affinity purified polyclonal antibody through custom antibody services at Covance Research Products Inc. (Denver, PA). Antisera from two rabbits was collected after an initial injection of 250 μg of purified D polypeptide in Freund’s complete adjuvant followed by three booster injections consisting of 125 μg of purified D polypeptide in incomplete Freund’s complete adjuvant. After determining antibody titer, the pooled sera was loaded onto a protein A Sepharose column. The affinity purified polyclonal anti-D antibody thus obtained was subsequently used in immunofluorescence and Western blot analysis.
HPIV 3 infection and preparation of cell extracts
HeLa cells (2 × 106) in a 100 mm dish were mock infected or infected with HPIV 3 at an MOI of 3 in OptiMEM (Invitrogen). At 2 h post-infection, cells were rinsed once with PBS and incubated for an additional 72 h in DMEM supplemented with 10% FBS. Cells were washed once in ice-cold PBS containing phosphatase inhibitors, scraped, and centrifuged at 2,000g for 5 min at 4°C to obtain a pellet. Total cell extracts were prepared following cell lysis with 500 μl of lysis buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% NP40) containing protease inhibitor cocktail (Sigma). For subcellular fractionation studies, cytoplasmic and nuclear extracts were isolated using the nuclear extract kit (Active Motif) according to manufacturer’s protocol. Briefly, after lysis in 500 μl of hypotonic buffer on ice for 15 min cells were centrifuged at 14,000g for 30 s at 4°C. The supernatant (cytoplasmic fraction) was carefully removed and the pellet was resuspended in 50 μl complete lysis buffer and incubated on ice for 30 min followed by centrifugation at 14,000g for 10 min at 4°C. The supernatant (nuclear extract) was removed. Protein concentration from cytoplasmic and nuclear extracts was determined and equal amount was separated on SDS-12% polyacrylamide gel for Western blot analysis.
Western blot analysis
Following electrophoresis, gels were electroblotted onto nitrocellulose membranes and blots were incubated for 1 h in blocking solution (5% skim milk and 0.02% Tween 20 in PBS). The blots were further incubated with the indicated primary antibodies, namely, polyclonal anti-His antibody (1:2,000) (SantaCruz), monoclonal anti-FLAG antibody (1:2,000) (Sigma), polyclonal anti-D antibody (1:15,000), monoclonal anti-actin antibody (1:2,000) (Santa Cruz), monoclonal anti-tubulin antibody (1:500) (Santa Cruz), and monoclonal anti-HN antibody (Fitzgerald) (1:1,000), in blocking solution for 1 h. Following incubation with the primary antibody, the blots were treated with the corresponding anti-mouse or anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Santa Cruz). After washing the blots three times in PBS-0.02% Tween 20, visualization was achieved using the ECL plus detection system (Optiblast) according to the manufacturer’s instruction.
Double-label immunofluorescence confocal microscopy
Subconfluent monolayers of HeLa cells (1.5 × 105) seeded onto coverslips in a six-well dish were mock infected or infected with HPIV 3 at an MOI of 3 in OptiMEM (Invitrogen). At 2 h post-infection, cells were rinsed once with PBS and incubated for 72 h in DMEM supplemented with 10% FBS. As indicated, HPIV 3 infected cells were additionally treated with 10 nM Leptomycin B (LMB) (Sigma) (LMB+) or DMSO alone (LMB−) for 4 h. Cells were fixed with 4% paraformaldehyde in PBS for 20 min, washed three times with PBS, and incubated with permeabilization solution (0.5% Triton X-100 in PBS) for 10 min. After being blocked for 1 h, cells were incubated individually in blocking buffer (3% normal goat serum, 0.1% Triton X-100 in PBS) containing polyclonal anti-D antibody (1:50,000) or simultaneously incubated in the presence of polyclonal anti-D antibody (1:50,000) and a monoclonal anti-HPIV 3 antibody (Fitzgerald) (1:1,000) overnight followed by individual or simultaneous incubations with anti-rabbit secondary antibody conjugated with Alexa 488 (Molecular Probes) (1:1,000) or anti-mouse secondary antibody conjugated to Alexa 568 (Molecular Probes) (1:1,000) for 2 h. After washing three times in PBS, cells were mounted in Vectashield (Molecular Probes) containing propidium iodide (PI) and observed under a Zeiss laser scanning microscope (Zeiss) using the appropriate excitation and emission wavelengths. Images were processed using the LSM imaging software (Zeiss).
Results
Alteration of HPIV 3 P gene and generation of PD ORF
The plasmid pGEM 4 Pflag was chosen for the manipulation of HPIV 3 P gene and successful generation of PD ORF was achieved in two steps. The insertion of two extra G residues within the “editing site” of P gene was achieved by QuikChange mutagenesis involving the use of QC1PD-F and QC2PD-R oligonucleotides (Table 1). Following mutagenesis, a second amplification reaction involving oligonucleotides, PD-ER5-F and PD-XH-R (Table 1), was undertaken and the modified EcoR V-Xho I fragment harboring two extra G residues was subsequently cloned into the corresponding sites of pGEM 4 Pflag to obtain pGEM 4 PDflag plasmid. The synthesis of full-length PD from pGEM 4 PDflag plasmid was finally verified by an in vitro transcription–translation coupled system (Promega) using [35S]-methionine as the labeled precursor following the manufacturer’s protocol. As can be seen from Fig. 1b, a 42 kDa band corresponding to the predicted size of full-length PD protein was readily transcribed and translated from rabbit reticulocytes incubated with pGEM 4 PDflag plasmid (lane 2). In addition, two minor polypeptides with a slightly lower molecular mass, presumably degradation products of full-length PD were also visible (Fig. 1b, lane 2). In contrast, no bands were observed from reticulocytes incubated with pGEM 4 plasmid used as empty vector control (Fig. 1b, lane 1). This result clearly demonstrated that synthesis of full-length PD could be accomplished using the in vitro system.
E. coli expression of full-length PD and the D polypeptide
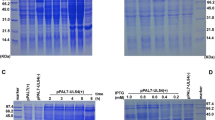
Our next goal was to express full-length PD as a His-tagged fusion protein using E. coli. To achieve this objective, DNA fragments encoding full-length PD and the 131 amino acids encompassing the D polypeptide, respectively, were cloned individually into plasmid pRSET-A. Following induction of E. coli BL21 (DE3) pLysS with 1 mM IPTG cell lysates were resolved on a 10% SDS-polyacrylamide gel and immunoblots were probed with anti-His antibody to reveal the presence of the recombinant proteins by ECL. As can be observed from Fig. 2a, bands corresponding to full-length PD (lane 2) and the D polypeptide (lane 4) were readily visible. Moreover, bands with identical sizes were also seen in the corresponding non-induced cell lysates indicating a basal level of protein expression (Fig. 2a, lanes 1 and 3). On the other hand, no bands could be detected in cell lysates containing the empty plasmid (Fig. 2a, lanes 5 and 6). Taken together, these results suggested that full-length PD as well as the D polypeptide could be expressed as bacterially His-tagged proteins.
(a). Bacterial expression of full-length PD and D polypeptides. E. coli BL21 (DE3) pLysS cells harboring pRSET-PD and pRSET-D plasmids were cultured in the presence (+) or absence (−) of IPTG to induce expression of the recombinant proteins. Cell lysates were analyzed by SDS-PAGE followed by Western blot with a polyclonal anti-His antibody. The presence of PD and D was ascertained after incubation of the blot with a secondary antibody conjugated to HRP followed by ECL detection. Cell lysate prepared from pRSET plasmid was used as a negative control. Arrows indicate the positions of PD and D. (b). Purification of D polypeptide by Ni-affinity column chromatography. Cells harboring pRSET-D plasmid were treated with IPTG to induce the expression of D polypeptide. Cell lysate prepared under denaturing conditions using 8 M urea was loaded onto a Ni-agarose column. Following elution with Tris-phosphate buffer containing 8 M urea over a pH gradient, fractions containing D polypeptide were analyzed by SDS-PAGE and visualized by Coomassie Brilliant Blue staining. The position of D polypeptide eluted under variable pH conditions: E1—5.9, E2 and E3—pH 4.5 are indicated by an arrow. (c). The IPTG induced pRSET-D and pRSET cell lysates from (a) were resolved by SDS-PAGE followed by Western blot analysis with affinity purified polyclonal anti-D antibody. The presence of D polypeptide was ascertained after incubation of the immunoblot with a secondary antibody conjugated to HRP followed by ECL detection. The position of D is denoted by an arrow
Partial purification of D polypeptide and immunoblot analysis with polyclonal D antibody
Since full-length PD comprises 241 amino acids from the P ORF fused to the 131 amino acids from the D ORF, we decided to partially purify the D polypeptide with an aim to generate a polyclonal D antibody. Accordingly, a large-scale (200 ml) E. coli BL21 (DE3) pLysS culture harboring pRSET-D plasmid was induced as mentioned before. Cell lysates were prepared under denaturing conditions in the presence of 8 M urea and loaded onto a Ni-agarose column and the His-tagged D polypeptide was eluted in the presence of 8 M urea over a pH gradient. Aliquots from each fraction were resolved by SDS-PAGE followed by staining with Coomassie Blue. As shown in Fig. 2b, efficient elution of His-tag D polypeptide was observed with a gradual lowering of pH (lanes 4 and 5) and a further reduction in the pH of elution buffer to 4.5 was not only seen to completely elute the protein but also increased its purity up to 90% as visualized by Coomassie staining (Fig. 2b, lane 6). The urea present in the partially purified protein fraction was removed by a desalting column and resuspended in PBS for subsequent generation of an affinity purified polyclonal antibody obtained after protein A Sepharose column chromatography.
The specificity of the affinity purified polyclonal anti-D antibody was further tested in a Western blot analysis using an E. coli lysate that expressed His tagged D polypeptide as described earlier. As shown in Fig. 2c, the antiserum was able to specifically recognize the D polypeptide (lane 2) while no reaction could be observed with the control E. coli lysate (Fig. 2c, lane 1) thereby confirming that the polyclonal anti-D antibody was able to specifically recognize the bacterially expressed protein. The polyclonal anti-D antibody was subsequently utilized in confocal microscopy analysis for the intracellular localization of PD in HPIV 3 infected cells.
Detection of full-length PD in vitro and in HPIV 3 infected cells in vivo
Since vaccinia virus/T7 RNA polymerase system is routinely utilized for transient gene expression in our laboratory, we wanted to determine whether transient expression of PD could be achieved using this approach. Accordingly, HeLa cells were infected with vaccinia virus vTF 7-3 and individually transfected with pGEM 4 Pflag and pGEM 4 PDflag plasmids as well as non-tagged pGEM 4 P and pGEM 4 PD plasmids as described and cell lysates were resolved on 10% SDS-polyacrylamide gels. Following electro transfer of proteins to nitrocellulose membrane, blots were individually probed with monoclonal anti-FLAG antibody or polyclonal anti-D antibody, washed three times in PBS-0.02% Tween 20, and further incubated with the corresponding horseradish peroxidase-labeled secondary antibody and detection with ECL. As expected, Western blot analysis with anti-Flag antibody successfully detected Pflag and PDflag proteins in cell extracts transfected with pGEM Pflag and pGEM 4 PDflag plasmids, respectively (Fig. 3a, lanes 2 and 4), while no bands were observed for the corresponding non-tagged P and PD proteins (Fig. 3a, lanes 3 and 5). Alternatively, incubation with polyclonal anti-D antibody successfully revealed the presence of PDflag and the non-tagged PD protein (Fig. 3b, lanes 4 and 5, respectively) and as predicted no reaction to either Pflag or P could be observed (Fig. 3b, lanes 2 and 3). In either case, no bands could be detected for the control pGEM 4 plasmid using anti-Flag or anti-D antibodies (Fig. 3a, b; lane 1).
(a). In vitro and in vivo detection of PD. Transient expression of PD in vitro was mediated by recombinant vaccinia virus/T7 RNA polymerase system. HeLa cells were infected with recombinant vaccinia virus vTF 7-3 that expresses T7 RNA polymerase at an MOI of 1 for 1 h and individually transfected with Flag-epitope tagged plasmids; pGEM 4Pflag and pGEM 4PDflag, as well as non-tagged pGEM 4P, pGEM 4PD, and control pGEM plasmids using Lipofectin according to manufacturer’s instruction. Equal amounts of cell lysates prepared 24 h post-transfection was analyzed by SDS-PAGE followed by a Western blot using a monoclonal anti-FLAG antibody or polyclonal anti-D antibody (b) and bands corresponding to Flag-tagged and non-tagged P and PD proteins were detected after incubation with an appropriate secondary antibody conjugated to HRP followed by ECL. Arrows indicate the position of tagged and non-tagged P and PD proteins in (a) and (b). (c). Detection of PD in HPIV 3 infected cells in vivo. HeLa cells were infected with HPIV 3 at an MOI of 3 and 72 h post-infection; total cell extracts were resolved by SDS-PAGE. Following electro transfer of proteins onto nitrocellulose membranes, Western blot was incubated with polyclonal anti-D (top), monoclonal anti-HN (middle), and monoclonal anti-actin (bottom) antibodies followed by incubation with the corresponding secondary antibodies conjugated to HRP and detection with ECL. Arrows indicate the positions of PD, HN, and actin
Detection of PD in HPIV 3 infected cells in vivo was achieved following SDS-PAGE and Western blot analysis as previously mentioned using polyclonal anti-D antibody. As anticipated, a band corresponding to the size of PD was apparent in cell extracts indicating that PD was indeed synthesized upon infection with HPIV 3 (Fig. 3c, top, lane 2). In addition, no other bands could be detected from infected cell extract suggesting the specificity of the polyclonal anti-D antibody to recognize PD. On the other hand, no bands were present in mock infected cell extracts (Fig. 3c, top, lane 1). Moreover, Western blot analysis of cell extracts with monoclonal anti-HN antibody additionally confirmed that cells were infected with HPIV 3 (Fig. 3c, middle, lane 2). Furthermore, cellular levels of actin, determined following incubation of Western blot with monoclonal anti-actin antibody, remained relatively constant (Fig. 3c, bottom, lanes 1 and 2). Conversely Western blot experiments utilizing an antibody directed toward the common NH2-terminus that is able to recognize both P and PD proteins could not be performed for the in vitro and in vivo detection of these proteins due to the lack of the availability of this specific antibody.
Taken together, results from Western blot analysis confirmed both the presence and specificity of the polyclonal anti-D antibody in recognizing PD in HPIV infected cells. Further studies involving immunofluorescence analysis were undertaken to ascertain the presence of PD protein in HPIV 3 infected cells.
Intracellular localization of PD in HPIV 3 infected cells
To examine the intracellular localization of PD, HeLa cells were infected with HPIV 3 at an MOI of 3 for 72 h, fixed with paraformaldehyde and permeabilized as mentioned. After incubation of cells with polyclonal anti-D antibody followed by a secondary antibody conjugated to Alexa 488, cells were viewed under a Zeiss microscope. A similar protocol was utilized for control mock infected HeLa cells. As shown in Fig. 4a, PD was uniformly distributed in the cytoplasm and also clearly visible within the nuclei of HPIV 3 infected cells (HPIV 3), the latter being apparent upon simultaneous staining of the cells with PI to reveal the presence of the nuclei. On the other hand, no reaction with the polyclonal anti-D antibody was observed with mock infected HeLa cells used as a control (Fig. 4a, Mock). To further confirm the intracellular localization of PD, equal amounts of cytoplasmic (C) and nuclear (N) fractions from mock infected (Mock) and HPIV 3 infected (HPIV 3) cells were resolved by SDS-PAGE and Western blots were probed with polyclonal anti-D and monoclonal anti-HN antibodies. As expected, PD was essentially present within the nuclear fraction while low levels were also observed in the cytoplasmic fraction of HPIV 3 infected cells (Fig. 4b, top, lanes 3 and 4). In addition, a band of about 70 kDa specific to HPIV 3 HN protein that was present within the cytoplasmic fraction confirmed that the cells were infected with HPIV 3 (Fig. 4b, middle, lane 3). As a positive control, a Western blot probed with anti-tubulin antibody demonstrated an equal amount of tubulin which is specifically present only in the cytoplasmic fraction (Fig. 4b, bottom, lanes 1 and 3). Taken together, these results suggested that PD was localized within both cytoplasmic and nuclear fractions of HPIV 3 infected cells.
(a) Localization of PD in HPIV 3 infected cells. Mock infected (Mock) and HPIV 3 infected HeLa cells (HPIV 3) were fixed and permeabilized as mentioned in section “Materials and methods” and incubated in the presence of polyclonal anti-D antibody. After incubation with a secondary antibody conjugated to Alexa 488, cells were mounted in Vectashield medium containing PI and viewed under a Zeiss fluorescence microscope. Images were acquired using Axiovert image processing software. The presence of PD within the nuclei of cells stained with PI is indicated by arrows. (b). Subcellular fractionation and intracellular localization of PD in HPIV 3 infected HeLa cells. Equal amounts of cytoplasmic (C) and nuclear (N) fractions isolated using nuclear extract kit (Active Motif) from mock infected (Mock) and infected (HPIV 3) cells were resolved by SDS-PAGE and Western blots were incubated with polyclonal anti-D (top), monoclonal anti-HN (middle), and monoclonal anti-tubulin (bottom) antibodies, respectively. The presence of PD, HN, and tubulin within each fraction was determined following incubation with the corresponding secondary antibodies conjugated to HRP and ECL. The position of individual proteins is denoted by an arrow
Double-labeled immunofluorescence confocal microscopy of PD in HPIV 3 infected cells
The precise localization of PD in HPIV 3 infected cells was determined by double-label immunofluorescence confocal microscopy analysis following Leptomycin B (LMB) treatment since several earlier reports have extensively utilized LMB as a tool to study the transport of proteins [32–37]. Accordingly, mock infected (Mock), or HPIV 3 infected cells, were non-treated (LMB−) or treated with LMB (LMB+) and simultaneously incubated with polyclonal anti-D antibody and a monoclonal anti-HPIV 3 antibody (Fitzgerald) followed by subsequent labeling with secondary antibodies conjugated to Alexa 488 and Alexa 568, respectively, were observed under a Zeiss confocal microscope. As can be seen in Fig. 5 (merge), an accumulation of PD in the nuclei and cytoplasm was evident in non-treated (LMB−) HPIV 3 infected cells along with other predominantly cytoplasmic viral antigen recognized by anti-HPIV 3 antibody (anti-HPIV 3) leading to the conclusion that PD was present both within the cytoplasm and nuclei of HPIV 3 infected cells. This nucleo-cytoplasmic pattern of PD observed in non-treated (LMB−) HeLa cells was altered upon treatment of HPIV 3 infected cells with LMB (LMB+). A significant amount of PD was now retained within the nuclei of HPIV 3 infected cells (compare merged images (LMB−) and (LMB+)) and its virtual absence from the cytoplasm was seen to enhance the staining of viral antigens by anti-HPIV 3 antibody (anti-HPIV 3) which was also evident in the merged image (see merged (LMB+)). In either case, no signals could be detected from the mock infected, non-treated (LMB−), or treated (LMB+) HeLa cells stained simultaneously with both anti-D and anti-HPIV 3 antibodies used as controls (Mock).
Double-labeled immunofluorescence confocal microscopy and co-localization of PD in HPIV 3 infected cells. HeLa cells were mock infected (Mock) or infected with HPIV 3 for 72 h and further treated with Leptomycin B (LMB+) or non-treated (LMB−). Cells were fixed, permeabilized, and double labeled with polyclonal anti-D antibody (anti-D) and monoclonal anti-HPIV 3 antibody (anti-HPIV 3) followed by secondary antibodies conjugated Alexa 488 (green) and Alexa 568 (red), respectively. Cells were observed under a Zeiss confocal immunofluorescence microscope and individual images acquired in appropriate wavelengths were generated and superimposed (merge) to reveal the co-localization of PD
In summary, results from cell fractionation studies together with confocal microscopy analysis following treatment with LMB provided conclusive evidence to demonstrate that the PD protein was present in both the cytoplasm and nuclei of HPIV 3 infected cells.
Discussion
Current studies to ascertain the presence of the PD protein were undertaken based on earlier reports indicating that full-length PD protein is likely to be synthesized from P mRNA via a transcriptional editing mechanism during HPIV 3 transcription [29] and a similar mechanism is also known to lead the synthesis of PD protein from bovine parainfluenza virus type 3 P mRNA [38]. Although the presence of PD in HPIV 3 infected cells has remained elusive, its importance in viral replication is borne by the fact that rDVKO virus devoid of both V and D ORF’s demonstrated significant degree of attenuation in vivo indicating that PD protein may play an important role in HPIV 3 replication in vitro, in vivo, or both [28].
In this report, we were able to successfully manipulate and subsequently clone the P mRNA that was fully capable of synthesizing the HPIV 3 PD protein in vitro. In addition, we also expressed both full-length PD and its unique COOH-terminal portion comprising 131 amino acid residues individually as His-tag fusion proteins for the generation of a polyclonal antibody enabling us to ascertain the presence and precise localization of PD in HPIV 3 infected cells by double-labeled immunofluorescence confocal microscopy revealing for the first time its presence predominantly within the nuclei and lower levels in the cytoplasm of HPIV 3 infected cells (Figs. 4 and 5). The ability of PD to localize between the cytoplasm and nucleus of infected cells suggests that it is likely to shuttle between these compartments as evidenced from our initial observation involving LMB treatment of HPIV 3 infected cells. Moreover, this localization of PD in HPIV 3 infected cells indicates that it may share some similarities with the Paramyxovirinae C and V proteins which have also been shown to localize between the cytoplasm and the nucleus of infected cells [39–43]. Furthermore, several nuclear localization signals with unique sequence configurations and capability to localize proteins to the nucleus have been successfully uncovered in a number of RNA viruses [44]. Surprisingly, a computational analysis of PD using PSORT II program [45] also predicted the presence of two motifs at positions, 266 PHQKGKR 272 and 340 PNPRHKR 346 within its COOH-terminus that complied with the ‘pat 7’ criteria for nuclear localization and comprising a proline residue followed within three residues by a basic segment containing three basic residues out of four [46]. Additionally, a third motif residing within residues, 225 KKSSSTHQEDDKRIKKG 241 was consistent with the bipartite nuclear localization signal constituting of two clusters of basic residues separated by a spacer of about 10 residues was similarly identified. It is fascinating to note that 225 KKSSSTHQEDDKRIKKG 241 sequence is located upstream of the “editing site” of P mRNA and is therefore also present within the P protein. Although, computational analysis demonstrated the presence of nuclear localization signals within PD, results from LMB treatment of HPIV 3 infected cells led to a significant retention of PD within the nuclei indicating that a nuclear export signal is also involved in the export of PD. In either case, a detailed study is necessary to unravel the mechanism of trafficking of PD. One possible line of inquiry currently under investigation involves mutational analysis of one or more of the consensus residues leading to the identification of nuclear targeting signals within PD. Another alternative possibility being considered in our laboratory involves fusion of the putative sequence to the enhanced green fluorescent protein to enable the ultimate intracellular localization of PD. It is tempting to speculate here that host cellular protein may play an important role in this process. One approach to resolve this issue shall utilize in vitro binding assays leading to the identification of the interacting protein and subsequent characterization of host responses to virus infections.
In summary, results from our initial study provide convincing evidence for presence and the intracellular localization of PD in HPIV 3 infected cells and assist us in pursuing further experiments aimed toward dissecting its role in HPIV 3 life cycle.
References
R.A. Lamb, D. Kolakofsky, in Field’s Virology, 4th edn. ed. by D.M. Knipe, P.M. Howley (Lippincott Williams and Wilkins, Philadelphia, 2001), pp. 1305–1340
R.M. Chanock, B.R. Murphy, P.L. Collins, in Field’s Virology, 4th edn. ed. by D.M. Knipe, P.M. Howley (Lippincott Williams and Wilkins, Philadelphia, 2001), pp. 1341–1379
A.K. Banerjee, S. Barik, B.P. De, Pharmacol. Ther. 51, 47–70 (1991). doi:10.1016/0163-7258(91)90041-J
B.P. De, A. Lesoon, A.K. Banerjee, J. Virol. 65, 3268–3275 (1991)
B.P. De, A.L. Burdsall, A.K. Banerjee, J. Biol. Chem. 268, 5703–5710 (1993)
B.P. De, A.K. Banerjee, Adv. Virus Res. 48, 169–204 (1997). doi:10.1016/S0065-3527(08)60288-2
B.P. De, S. Gupta, S. Gupta, A.K. Banerjee, Proc. Natl. Acad. Sci. USA. 92, 5204–5208 (1995). doi:10.1073/pnas.92.11.5204
B.P. De, M.A. Hoffman, S. Choudhary, C.C. Huntley, A.K. Banerjee, J. Virol. 74, 5886–5895 (2000). doi:10.1128/JVI.74.13.5886-5895.2000
C.C. Huntley, B.P. De, N.R. Murray, A.P. Fields, A.K. Banerjee, Virology 211, 561–567 (1995). doi:10.1006/viro.1995.1438
S.K. Choudhary, A.G. Malur, Y. Huo, B.P. De, A.K. Banerjee, Virology 302, 373–382 (2002). doi:10.1006/viro.2002.1668
H. Zhao, A.K. Banerjee, J. Biol. Chem. 270, 12485–12490 (1995). doi:10.1074/jbc.270.21.12485
M.S. Galinski, M.A. Mink, D.M. Lambert, S.L. Wechsler, M.W. Pons, Virology 154, 46–60 (1986). doi:10.1016/0042-6822(86)90167-4
D. Luk, A. Sanchez, A.K. Banerjee, Virology 153, 318–325 (1986). doi:10.1016/0042-6822(86)90036-X
M.K. Spriggs, P.L. Collins, J. Gen. Virol. 67, 2705–2719 (1986)
J. Curran, J.B. Marq, D. Kolakofsky, Virology 189, 647–656 (1992). doi:10.1016/0042-6822(92)90588-G
T. Cadd, D. Garcin, C. Tapparel, M. Itoh, M. Homma, L. Roux et al., J. Virol. 70, 5067–5074 (1996)
C. Tapparel, S. Hausmann, T. Pelet, J. Curran, D. Kolakofsky, L. Roux, J. Virol. 71, 9588–9599 (1997)
S.M. Horikami, R.M. Hector, S. Smallwood, S.A. Moyer, Virology 235, 261–270 (1997). doi:10.1006/viro.1997.8702
D.A. Sweetman, J. Miskin, M.D. Baron, Virology 281, 193–204 (2001). doi:10.1006/viro.2000.0805
G.C. Grogan, S.A. Moyer, Virology 288, 96–108 (2001). doi:10.1006/viro.2001.1068
S. Smallwood, S.A. Moyer, Virology 318, 439–450 (2004). doi:10.1016/j.virol.2003.09.045
B. Bankamp, J. Wilson, W.J. Bellini, P.A. Rota, Virology 336, 120–129 (2005). doi:10.1016/j.virol.2005.03.009
A.G. Malur, M.A. Hoffman, A.K. Banerjee, Virus Res. 99, 199–204 (2004). doi:10.1016/j.virusres.2003.11.009
A.G. Malur, S. Chattopadhyay, R.K. Maitra, A.K. Banerjee, J. Virol. 79, 7877–7882 (2005). doi:10.1128/JVI.79.12.7877-7882.2005
B. Gotoh, T. Komatsu, K. Takeuchi, J. Yokoo, Microbiol. Immunol. 45, 787–800 (2001)
B. Gotoh, T. Komatsu, K. Takeuchi, J. Yokoo, Rev. Med. Virol. 12, 337–357 (2002). doi:10.1002/rmv.357
S. Bose, A.K. Banerjee, J. Interferon Cytokine Res. 3, 401–412 (2003). doi:10.1089/107999003322277810
A.P. Durbin, J.M. McAuliffe, P.L. Collins, B.R. Murphy, Virology 261, 319–330 (1999). doi:10.1006/viro.1999.9878
M.S. Galinski, R.M. Troy, A.K. Banerjee, Virology 186, 543–555 (1992). doi:10.1016/0042-6822(92)90020-P
A. Bukreyev, M.H. Skiadopoulos, B.R. Murphy, P.L. Collins, J. Virol. 70, 10293–10306 (2003)
A.G. Malur, S.K. Choudhary, B.P. De, A.K. Banerjee, J. Virol. 76, 8101–8109 (2002). doi:10.1128/JVI.76.16.8101-8109.2002
M. Fukuda, S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida et al., Nature 390, 308–311 (1997). doi:10.1038/36894
B. Wolff, J.J. Sanglier, Y. Wang, Chem. Biol. 4, 139–147 (1997). doi:10.1016/S1074-5521(97)90257-X
N. Kudo, B. Wolff, T. Sekimoto, E.P. Shreiner, Y. Yoneda, M. Yanagida et al., Exp. Cell Res. 242, 540–547 (1998). doi:10.1006/excr.1998.4136
N. Kudo, N. Matsumori, H. Taoka, D. Fujiwara, E.P. Shreiner, B. Wolff et al., Proc. Natl. Acad. Sci. USA. 96, 9112–9117 (1999). doi:10.1073/pnas.96.16.9112
D. Pasdeloup, N. Poisson, H. Raux, Y. Gaudin, R.W.H. Ruigrok, D. Blondel, Virology 334, 284–293 (2005). doi:10.1016/j.virol.2005.02.005
H. Sato, M. Masuda, R. Miura, M. Yoneda, C. Kai, Virology 352, 121–130 (2006). doi:10.1016/j.virol.2006.04.013
T. Pelet, J. Curran, D. Kolakofsky, EMBO J. 10, 443–448 (1991)
W.J. Bellini, G. Englund, S. Rozenblatt, H. Arnheiter, C.D. Richardson, J. Virol. 53, 908–919 (1985)
G. Alkhatib, B. Massie, D.J. Briedis, J. Virol. 62, 4059–4069 (1988)
N. Watanabe, M. Kawano, M. Tsurudome, S. Kusagawa, M. Nishio, H. Komada, T. Shima, Y. Ito, J. Gen. Virol. 77, 327–338 (1996)
M. Nishio, M. Tsurudome, M. Ito, M. Kawano, S. Kusugawa, H. Komada, Y. Ito, Med. Microbiol. Immunol. 188, 79–82 (1999)
T. Nishie, K. Nagata, K. Takeuchi, Microbes Infect. 9, 344–354 (2007)
J.A. Hiscox, Virus Res. 95, 13–22 (2003)
K. Nakai, P. Horton, Trends Biochem. Sci. 24, 34–36 (1999)
I. Macara, Microbiol. Mol. Biol. Rev. 65, 570–594 (2001)
Acknowledgments
This work was supported by North Carolina State New Investigator’s Research Award to A.G.M.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wells, G., Malur, A. Expression of human parainfluenza virus type 3 PD protein and intracellular localization in virus infected cells. Virus Genes 37, 358–367 (2008). https://doi.org/10.1007/s11262-008-0269-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-008-0269-2